Józef Adam Liwo
List of publications and invited presentations
Original papers in ISI journals
- J. Kostrowicki, A. Liwo. DECFAM - A new computer-oriented
algorithm for the determination of equilibrium constants from potentiometric
and/or spectrophotometric measurements - I. Comput. Chem. 1984, 8,
91-99.
- J. Kostrowicki, A. Liwo. DECFAM - A new computer-oriented
algorithm for the determination of equilibrium constants from potentiometric
and/or spectrophotometric measurements - II. Comput. Chem. 1984, 8,
101-105.
- A. Liwo, J. Ciarkowski. Origin of the ring-ring interaction in
cyclic dipeptides incorporating an aromatic amino acid. Tetrahedron Lett.
1985, 26, 1873-1876.
- Z. Warnke, C. Trojanowska, A. Liwo. Potentiometric study on the
complex formation of some divalent metal ions with 2-aminoxyacids.
J. Coord. Chem. 1985, 14, 31-38.
- L. Chmurzyński, A. Liwo, A. Wawrzynów, A. Tempczyk.
Theoretical and experimental studies on the UV spectra of pyridine N-oxide
perchlorates. J. Mol. Struct. 1986, 143, 375-378.
- A. Wawrzynów, A. Liwo, L. Chmurzyński, A. Tempczyk.
A study on the UV spectra of pyridine N-oxide and 2,6-lutidine N-oxide
1:1 perchlorates solutions in propylene carbonate. J. Mol. Struct.
1986, 143, 379-382.
- J. Kostrowicki, A. Liwo. A general method for the determination
of the stoichiometry of unknown species in multicomponent systems from
physicochemical measurements. Comput. Chem. 1987, 11, 195-210.
- J. Kostrowicki, A. Liwo, K. Sokołowski. A comparative study
on some methods for computing of equilibrium concentrations. Comput. Chem.
1988, 12, 293-299.
- A. Wawrzynów, K. Sokołowski, A. Liwo, L. Chmurzyński.
Experimental studies on the UV spectra of substituted pyridine N-oxides.
J. Mol. Struct. 1988, 174, 235-240.
- A. Liwo, A. Tempczyk, Z. Grzonka. Molecular mechanics
calculations on deaminooxytocin and on deamino arginine vasopressin and its
analogues. J. Comput.-Aided Mol. Design 1988, 2, 281-309.
- A. Liwo, J. Ciarkowski. Conformation of dioxopiperazines - 8.
The model building algorithm. J. Mol. Struct. THEOCHEM 1989,
183, 331-340.
- A. Liwo, A. Tempczyk, Z. Grzonka. Theoretical studies of the
mechanism of the action of the neurohypophyseal hormones. I. Molecular
electrostatic potential (MEP) and molecular electrostatic field (MEF) maps of
some vasopressin analogues. J. Comput.-Aided Mol. Design 1989, 3,
261-284.
- A. Tempczyk, M. Tarnowska, A. Liwo. A theoretical study of
glucosamine synthase. I. Molecular mechanics calculations on substrate binding.
Eur. Biophys. J. 1989, 17, 201-210.
- L. Chmurzyński, A. Liwo, A. Tempczyk. Relationship between
the electronic structure and acidic-basic properties of some substituted
pyridine N-oxides. Z. Naturforsch. B 1989, 44b, 1263-1270.
- L. Chmurzyński, A. Liwo. Influence of the conjugation effect
on the UV-spectra of 4-substituted pyridine N-oxides and their conjugated
acids. J. Mol. Struct. 1990, 218, 129-134.
- J. Kostrowicki, A. Liwo. Determination of equilibrium parameters
by minimization of an extended sum of squares. Talanta 1990, 37,
645-650.
- L. Chmurzyński, A. Liwo. A CNDO/2 study of the homoconjugation
energies of 4-substituted pyridine N-oxides. Z. Naturforsch. B 1990,
45b, 717-718.
- L. Chmurzyński, A. Liwo, A. Wawrzynów, A. Tempczyk.
CNDO/S-CI-nPDQ studies of the solvation effect on the UV-spectra of pyridine
N-oxide and its complexes with proton. Z. Naturforsch. B 1990,
45b, 719-720.
- A. Liwo, K. Sokołowski, A. Wawrzynów, L. Chmurzyński.
UV-spectroscopic studies on the influence of the traces of water on the
protolytic equilibria of substituted pyridine N-oxides in aprotic solvents.
J. Sol. Chem. 1990, 19, 1113-1124.
- Z. Pawlak, S. Kuna, M. Richert, E. Giersz, M. Wiśniewska, A. Liwo,
L. Chmurzyński. Acidity constants of 19 protonated N-bases in
cyclohexanone, acetone, and butan-2-one. J. Chem. Thermodyn. 1991,
23, 135-140.
- A. Liwo, T. Widernik, T. Klentak. Influence of inorganic anions
on the tautomeric equilibria of N-substituted aminoazobenzenes - the evidence
for ionic association. J. Sol. Chem. 1991, 20, 431-443.
- L. Chmurzyński, A. Liwo. Simple methods for the estimation
of ionization constants of substituted pyridine N-oxides in polar aprotic
solvents and water. J. Sol. Chem. 1991 20, 731-738.
- M.D. Shenderovich, F. Kasprzykowski, A. Liwo, I. Sekacis,
J. Saulitis, G.V. Nikiforovich. Conformational analysis of [Cpp
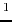 ,Sar
,Sar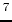 ,
Arg
,
Arg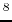 ]vasopressin by
]vasopressin by 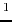 H-NMR spectroscopy and molecular mechanics
calculations. Int. J. Pept. Protein Res. 1991, 38, 528-538.
H-NMR spectroscopy and molecular mechanics
calculations. Int. J. Pept. Protein Res. 1991, 38, 528-538.
- L. Chmurzyński, A. Liwo. Acid-base and cationic homoconjugation
equilibria of substituted pyridine N-oxides in acetone. J. Chem. Soc.
Faraday Trans. 1991, 87, 3853-3856.
- J. Tarasiuk, A. Liwo, W. Wojtkowiak, M. Dzieduszycka, A. Tempczyk,
A. Garnier-Suillerot, S. Martelli, E. Borowski.
Molecular determinants of singlet oxygen binding by anthraquinones in relation
to their redox cycling activity.
Anti-Cancer Drug Design 1991, 6, 399-416.
- A. Tempczyk, M. Tarnowska, A. Liwo, E. Borowski.
A theoretical study of glucosamine synthase. II. Combined quantum and molecular
mechanics simulation of sulfhydryl attack on the carboxyamide group.
Eur. Biophys. J., 1992, 21, 137-145.
- A. Liwo, M. Tarnowska, Z. Grzonka, A. Tempczyk. Modified
Free-Wilson method for the analysis of biological activity data.
Comput. Chem. 1992, 16, 1-9.
- M.H. Hao, S. Rackovsky, A. Liwo, M.R. Pincus, H.A. Scheraga.
Effects of compact volume and chain stiffness on the conformations of native
proteins.
Proc. Natl. Acad. Sci. U.S.A., 1992, 89, 6614-6618.
- M. Tarnowska, S. Ołdziej, A. Liwo, P. Kania, F. Kasprzykowski,
Z. Grzonka. MNDO study of the mechanism of the inhibition of cysteine
proteinases by diazomethyl ketones.
Eur. Biophys. J., 1992, 21, 217-222.
- M. Tarnowska, S. Ołdziej, A. Liwo, Z. Grzonka, E. Borowski.
Investigation of the inhibition pathway of glucosamine synthase by
N
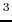 -(4-methoxyfumaroyl)-L-2,3-diaminopropanoic acid by semiempirical quantum
mechanical and molecular mechanics methods.
Eur. Biophys. J., 1992, 21, 273-280.
-(4-methoxyfumaroyl)-L-2,3-diaminopropanoic acid by semiempirical quantum
mechanical and molecular mechanics methods.
Eur. Biophys. J., 1992, 21, 273-280.
- D. Jeziorek, D. Dyl, A. Liwo, W. Woźnicki, A. Tempczyk, E. Borowski.
A theoretical study of the mechamism of oxygen binding by model anthraquinones
I: Quantum mechanical evaluation of the oxygen-binding sites of
1,4-hydroquinone.
Anti-Cancer Drug Design, 1992, 7, 451-461.
- D. Jeziorek, D. Dyl, A. Liwo, W. Woźnicki, A. Tempczyk, E. Borowski.
A theoretical study of the mechamism of oxygen binding by model anthraquinones
II: Quantum mechanical studies of the energetics of oxygen binding
to model anthraquinones.
Anti-Cancer Drug Design, 1993, 8, 223-235.
- A. Liwo, M.R. Pincus, R.J. Wawak, S. Rackovsky, H.A. Scheraga.
Calculation of protein backbone geometry from
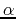 -carbon coordinates
based on peptide-group dipole alignment.
Protein Science, 1993, 2, 1697-1714.
-carbon coordinates
based on peptide-group dipole alignment.
Protein Science, 1993, 2, 1697-1714.
- A. Liwo, M.R. Pincus, R.J. Wawak, S. Rackovsky, H.A. Scheraga.
Prediction of protein conformation on the basis of a search for compact
structures; test on avian pancreatic polypeptide.
Protein Science, 1993, 2, 1715-1731.
- M. Tarnowska, A. Liwo, M.D. Shenderovich, I. Liepiņa,
A.A. Golbraikh, Z. Grzonka, A. Tempczyk.
A molecular mechanics study of the effect of substitution in position 1 on the
conformational space of the oxytocin/vasopressin ring.
J. Comput.-Aided Mol. Design, 1993, 7, 699-719.
- A. Liwo, A. Tempczyk, T. Widernik, T. Klentak, J. Czermiński.
A theoretical study of protonation and tautomerization of N-substituted
aminoazobenzenes.
J. Chem. Soc. Perkin Trans. 2, 1994, 71-75.
- J. Ciarkowski, St. Ołdziej, A. Liwo.
Mimicking the mechanism of aspartic proteases. Part 1. Molecular mechanics study.
Pol. J. Chem., 1994, 68, 939-947.
- J. Ciarkowski, A. Liwo, S. Ołdziej, M. Shenderovich.
Oxytocin antagonists' pharmacophore.
Pol. J. Chem., 1994, 68, 929-938.
- M. Kawińska, L. Łankiewicz, P. Skurski, B. Micewicz, W. Wiczk
St. Ołdziej, A. Liwo.
Fluorescence energy transfer in a series of leucine-enkephalin analogues.
Pol. J. Chem., 1994, 68, 975-985.
- F. Kasprzykowski, P. Skurski, A. Liwo, L. Łankiewicz S. Ołdziej
J. Łanoszka, W. Wiczk, Z.Grzonka.
Conformational studies of oxytocin analogues.
Pol. J. Chem., 1994, 68, 987-995.
- M. Shenderovich, W. Wilke, K. Kövér, N. Collins, V. Hruby,
A. Liwo, J. Ciarkowski.
Solution conformation of a potent bicyclic antagonist of oxytocin.
Pol. J. Chem., 1994, 68, 921-927.
- A. Liwo, K.D. Gibson, H.A. Scheraga, P.W. Brandt-Rauf, R. Monaco, M.R. Pincus.
Comparison of the low energy conformations of an oncogenic and a non-oncogenic
p21 protein, neither of which binds GTP or GDP.
J. Prot. Chem., 1994, 13, 237-251.
- A. Liwo, S. Ołdziej, J. Ciarkowski, G. Kupryszewski,
M.R. Pincus, R.J. Wawak, S. Rackovsky, H.A. Scheraga.
Prediction of conformation of rat galanin in the presence and absence of water
with the use of Monte Carlo methods and the ECEPP/3 force field.
J. Prot. Chem., 1994, 13, 375-380.
- Z. Pawlak, S. Kuna, M. Richert, E. Giersz, A. Liwo, L. Chmurzyński.
Proton transfer and heteroconjugation of ammonium ions with N-bases
in cyclohexanone, propanone, and butan-2-one.
J. Chem. Thermodynamics, 1994, 26, 483-492.
- A. Angielski, A. Liwo, D. Konopinska, J. Ciarkowski.
Theoretical Conformational analysis of insect neuropeptide proctolin and its
selected analogues.
Bull. Pol. Chem. Soc. Chemistry, 1994, 42, 268-273.
- D. Jeziorek, D. Dyl, A. Liwo, T. Ossowski, W. Woźnicki.
Enthalpy of oxygen addition to anthraquinone derivatives
determines their ability to mediating NADH oxidation.
Anti-Cancer Drug Design, 1994, 9, 435-448.
- J. Rak, P. Skurski, A. Liwo, J. Błażejowski.
Theoretical studies of the structure, stability, ability to internal
transformation and tautomerization, as well as reactivity of H
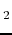 PPH
PPH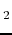 and HPPH
and HPPH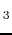 molecules.
J. Am. Chem. Soc., 1995, 117, 2638-2648.
molecules.
J. Am. Chem. Soc., 1995, 117, 2638-2648.
- S. Ołdziej, J. Ciarkowski, A. Liwo, M.D. Shenderovich, Z. Grzonka.
Conformational aspects of differences in requirements of oxytocin and
vasopressin receptors.
Journal of Receptor & Signal Transduction Research, 1995, 15,
703-713.
- R. Kaźmierkiewicz, A. Liwo, B. Lammek.
Theoretical conformational analysis of three vasopressin antagonists with a
modified cyclohexyl ring in the first thioacid residue.
Int. J. Pept. Prot. Res., 1995, 45, 451-458.
- D.R. Ripoll, M.S. Pottle, K.D. Gibson, A. Liwo, H.A. Scheraga.
Implementation of the ECEPP algorithm, the Monte Carlo Minimization method,
and the Electrostatically Driven Monte Carlo method on the Kendall Square
research KSR1 computer.
J. Comput. Chem., 1995, 16, 1153-1163.
- A. Liwo, D. Jeziorek, T. Ossowski, D. Dyl, A. Tempczyk, J. Tarasiuk,
M. Nowacka, E. Borowski, W. Woźnicki.
Molecular modeling of singlet-oxygen binding to
anthraquinones in relation to the
peroxidating activity of antitumor anthraquinone drugs.
Acta Biochim. Pol., 1995, 42, 445-456.
- A. Liwo, A. Tempczyk, S. Ołdziej, M.D. Shenderovich,
V.J. Hruby, S. Talluri, J. Ciarkowski, F. Kasprzykowski,
L. Łankiewicz, Zbigniew Grzonka.
Exploration of the Conformational Space of Oxytocin and
Arginine-Vasopressin
Using the Electrostatically-Driven Monte Carlo and Molecular Dynamics
Methods.
Biopolymers, 1996, 38, 157-175.
- W. Wiczk, P. Rekowski, G. Kupryszewski, J. Łubkowski,
S. Ołdziej, A. Liwo.
Fluorescence and Monte Carlo conformational studies of the (1-15)galanin
amide fragment.
Biophys. Chem., 1996, 58, 303-312.
- R.J. Wawak, K.D. Gibson, A. Liwo, H.A. Scheraga.
Theoretical prediction of a crystal structure.
Proc. Natl. Acad. Sci. USA, 1996, 93, 1743-1746.
- D.R. Ripoll, Yu. Vorobjev, A. Liwo, J.A. Vila, H.A. Scheraga.
Coupling between folding and ionization equilibrium. Effects of pH on the
conformational preferences of oligopeptides.
J. Mol. Biol., 1996, 264, 770-783.
- W. Wiczk, L. Łankiewicz, C. Czaplewski, S. Ołdziej, K.
Stachowiak, A. Michniewicz, A. Liwo.
The photophysics of
 -homotyrosine and its simple derivatives.
J. Photochem. Photobiol. A: Chemistry, 1996, 101, 171-181.
-homotyrosine and its simple derivatives.
J. Photochem. Photobiol. A: Chemistry, 1996, 101, 171-181.
- A. Liwo, P. Skurski, St. Ołdziej,
L. Łankiewicz, J. Malicka, W. Wiczk.
A new approach to the resolution of the excitation-emission
spectra of multicomponent systems.
Comput. Chem., 1997, 21, 89-96.
- D. Jeziorek, T. Ossowski, A. Liwo, D. Dyl, M. Nowacka, W. Woźnicki.
Theoretical and electrochemical study of the mechanism of
anthraquinone-mediated one-electron reduction of oxygen:
the involvement of adducts of dioxygen species to anthraquinones.
J. Chem. Soc. Perkin Transactions 2, 1997, 229-236.
- A. Liwo, St. Ołdziej, M.R. Pincus, R.J. Wawak, S. Rackovsky, H.A. Scheraga.
A united-residue force field for off-lattice protein-structure simulations.
I: Functional forms and parameters of long-range side-chain
interaction potentials from protein crystal data.
J. Comput. Chem., 1997, 18, 849-873.
- A. Liwo, M.R. Pincus, R.J. Wawak, S. Rackovsky, St. Ołdziej, H.A. Scheraga.
A united-residue force field for off-lattice protein-structure simulations.
II: Parameterization of local interactions and determination
of the weights of energy terms by Z-score optimization.
J. Comput. Chem., 1997, 18, 874-887.
- A. Liwo, D. Jeziorek, D. Dyl, T. Ossowski, L. Chmurzyński.
Theoretical Study of the role of hydrogen bonding and proton
transfer in oxygen reduction by semiquinones.
J. Mol. Struct. THEOCHEM, 1997, 388-389, 445-449.
- L. Chmurzyński, A. Liwo, P. Barczyński.
A potentiometric study of acid-base equilibria of substituted pyridine N-oxides
in nitrobenzene.
Anal. Chim. Acta, 1996, 335, 147-153.
- L. Chmurzyński, E. Kaczmarczyk, A. Liwo.
A study of cationic heteroconjugation equilibria of substituted pyridine
N-oxides in acetonitrile.
Anal. Chim. Acta, 1997, 338, 261-267.
- A. Liwo, D. Dyl, D. Jeziorek, M. Nowacka, T. Ossowski, W. Woźnicki.
MCSCF study of singlet-oxygen addition to ethenol - a model
of photooxidation reactions of unsaturated and aromatic compounds bearing
hydroxy groups.
J. Comput. Chem., 1997, 18, 1668-1681.
- W. Wiczk, L. Łankiewicz, C. Czaplewski, S. Ołdziej, K.
Stachowiak, A. Michniewicz, B. Micewicz, A. Liwo.
The photophysics of tyrosine and its simple derivatives.
J. Fluorescence, 1997, 7, 257-266.
- I. Liepiņa, M. Blanco, G. Duburs, A. Liwo.
Spatial structure of dihydropyridines and similarity of dihydropyridines
with some amino acids.
Molecular Engineering, 1997, 7, 401-427.
- A. Liwo, R. Kaźmierkiewicz, C. Czaplewski, M. Groth, S. Ołdziej,
R.J. Wawak, S. Rackovsky, M.R. Pincus, H.A. Scheraga.
United-residue force field for off-lattice protein-structure
simulations. III. Origin of backbone hydrogen-bonding cooperativity in
united-residue potentials, J. Comput. Chem., 1998, 19, 259-276.
- A. Liwo, D. Jeziorek, T. Ossowski, M. Groth.
CASSCF/MCQDPT2 study on electron and hydrogen-atom transfer from ethene
derivatives to singlet oxygen - implication for peroxidation processes,
Internet Journal of Chemistry, 1998 1, Article 13.
- X.W. Shi, A.B. Attygalle, A. Liwo, M.-H. Hao, J. Meinwald, H.R.W. Dharmaratne,
W.M.A.P. Wanigaseker.
Absolute stereochemistry of soulattrolide and its analogues.
J. Org. Chem., 1998, 63, 1233-1238.
- J. Malicka, M. Groth, C. Czaplewski, A. Liwo, W. Wiczk, L. Łankiewicz.
Computer modeling of the solution conformation of cyclic enkephalins,
Lett. Pept. Sci., 1998, 5, 445-447.
- A. Wawrzynów, A. Liwo, E. Kaczmarczyk, L. Chmurzyński.
A potentiometric study of the (OHO)
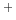 -type cationic heteroconjugation
equilibria in propylene carbonate.
Journal of Molecular Structure, 1998, 448, 185-189.
-type cationic heteroconjugation
equilibria in propylene carbonate.
Journal of Molecular Structure, 1998, 448, 185-189.
- A. Wawrzynów, A. Liwo, E. Kaczmarczyk, L. Chmurzyński.
Cationic heteroconjugation constants in the OHN
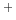 and NHO
and NHO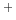 systems in
propylene carbonate: dependence on difference in basicity.
J. Solution Chem., 1998, 27, 463-472.
systems in
propylene carbonate: dependence on difference in basicity.
J. Solution Chem., 1998, 27, 463-472.
- R.J. Wawak, J. Pillardy, A. Liwo, K.D. Gibson, H.A. Scheraga.
Diffusion equation and distance scaling methods of global optimization:
applications to crystal structure prediction.
J. Phys. Chem. A, 1998, 102, 2904-2918.
- D.R. Ripoll, A. Liwo, H.A. Scheraga.
New developments of the Electrostatically Driven Monte Carlo Method: test
on the membrane-bound portion of melittin.
Biopolymers, 1998, 46, 117-126.
- A. Liwo, J. Pillardy, R. Kaźmierkiewicz, R.J. Wawak, M. Groth,
C. Czaplewski, S. Ołdziej, H.A. Scheraga.
Prediction of protein structure using a knowledge-based
off-lattice united-residue force field and global optimization methods.
Theoretical Chemistry Accounts, 1999, 101, 16-20.
- J. Lee, A. Liwo, H.A. Scheraga.
Energy-based de novo protein folding by conformational space
annealing and an off-lattice united-residue force field:
Application to the 10-55 fragment of staphylococcal protein A
and to apo calbindin D9K.
Proc. Natl. Acad. Sci. USA, 1999, 96, 2025-2030.
- E. Kaczmarczyk, R. Wróbel, A. Liwo, L. Chmurzyński.
Temperature dependence of the acid-base equilibrium constants of substituted
pyridine N-oxides in acetonitrile.
J. Mol. Struct., 1999, 477, 113-118.
- A. Liwo, J. Lee, D.R. Ripoll, J. Pillardy, H.A. Scheraga.
Protein structure prediction by global optimization
of a potential energy function.
Proc. Natl. Acad. Sci. USA, 1999, 96, 5482-5485.
- J. Lee, A. Liwo, D.R. Ripoll, J. Pillardy, H.A. Scheraga.
Calculation of Protein Conformation by Global Optimization
of a potential energy function.
Proteins: Struct. Func. Genet., 1999, Suppl. 3, 204-208.
- J. Pillardy, A. Liwo, M. Groth, H.A. Scheraga.
An efficient deformation-based global optimization method for off-lattice
polymer chains; self-consistent basin-to-deformed-basin mapping (SCBDBM).
Application to united-residue polypeptide chains.
J. Phys. Chem. B, 1999, 103, 7353-7366.
- J. Zielińska, M. Makowski, K. Maj, A. Liwo, L. Chmurzyński.
Acid-base and hydrogen-bonding equilibria in aliphatic amine
and carboxylic acid systems in non-aqueous solutions.
Anal. Chim. Acta, 1999, 401, 317-321.
- A. Bogdańska, L. Chmurzyński, T. Ossowski, A. Liwo, D. Jeziorek.
Protolytic equilibria of dihydroxyanthraquinones in non-aqueous solutions.
Anal. Chim. Acta, 1999, 402, 339-343.
- J. Pillardy, A. Liwo, H.A. Scheraga.
An efficient deformation-based global optimization method [self-consistent
basin-to-deformed-basin mapping (SCBDBM)]; Application to Lennard-Jones
atomic clusters.
J. Phys. Chem. A, 1999, 103, 9370-9377.
- M. Makowski, A. Liwo, R. Wróbel, L. Chmurzyński.
Ab initio study of the energetics of protonation
and hydrogen bonding of pyridine N-Oxide and its derivatives
J. Phys. Chem. A, 1999, 103, 11104-11108.
- S. Rodziewicz, I. Wirkus-Romanowska, M. Ciurak, H. Miecznikowska, G.
Kupryszewski, C. Czaplewski, A. Liwo, K. Rolka.
Cyclic analogues of proline-rich protein fragments. Part II:
Conformational studies using NMR spectroscopy and theoretical
conformational analysis.
Pol. J. Chem., 1999, 73, 1697-1710.
- J. Malicka, M. Groth, C. Czaplewski, A. Liwo, W. Wiczk.
Fluorescence decay time distribution analysis of cyclic enkephalin analogues.
Influence of the solvents and configuration of amino acids in position 2
and 3 on changes in conformation.
Acta Biochim. Pol., 1999, 46, 615-629.
- M. Groth, J. Malicka, C. Czaplewski, S. Ołdziej, L. Łankiewicz,
W. Wiczk, A. Liwo.
Maximum entropy approach to the determination of solution
conformation of
flexible polypeptides by global conformational analysis and
NMR spectroscopy - application to
DNS
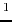 -c-[D-A
-c-[D-A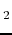 bu
bu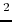 ,Trp
,Trp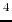 ,Leu
,Leu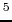 ]enkephalin
and DNS
]enkephalin
and DNS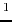 -c-[D-A
-c-[D-A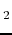 bu
bu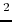 , Trp
, Trp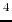 , D-Leu
, D-Leu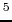 ]enkephalin.
J. Biomol. NMR, 1999, 4, 315-330.
]enkephalin.
J. Biomol. NMR, 1999, 4, 315-330.
- J. Lee, A. Liwo, D.R. Ripoll, J. Pillardy, J.A. Saunders, K.D. Gibson,
H.A. Scheraga.
Hierarchical energy-based approach to protein-structure prediction;
blind-test evaluation with CASP3 targets.
Int. J. Quant. Chem., 2000, 77, 90-117.
- J. Lee, J. Pillardy, C. Czaplewski, Y. Arnautova, D.R. Ripoll, A. Liwo,
K.D. Gibson, R.J. Wawak, H.A. Scheraga.
Efficient parallel algorithms in global optimization
of potential energy functions.
Comput. Phys. Comm., 2000, 128, 399-411.
- T. Ossowski, P. Pipka, A. Liwo, D. Jeziorek.
Electrochemical and UV-spectrophotometric study of oxygen and superoxide anion
radical interaction with anthraquinone derivatives and their radical anions.
Electrochim. Acta, 2000, 45, 3581-3587.
- C. Czaplewski, S. Rodziewicz-Motowidło, A. Liwo, D.R. Ripoll,
R.J. Wawak, H.A. Scheraga,
Molecular simulation study of cooperativity in hydrophobic association.
Prot. Sci., 2000, 9, 1235-1245.
- M. Bobrowski, A. Liwo, S. Ołdziej, D. Jeziorek, T. Ossowski,
CAS MCSCF/CAS MCQDPT2 study of the mechanism of
singlet-oxygen addition to 1,3-butadiene and benzene,
J. Am. Chem. Soc., 2000, 122, 8112-8119.
- S. Rodziewicz-Motowidło, A. Łęgowska, X.F. Qi, C. Czaplewski, A. Liwo,
P. Sowiński, W. Mozga, J. Olczak, J. Zabrocki, K. Rolka.
Solution conformational study of Scyliorhinin I analogues with conformational
constraints by two-dimensional NMR and theoretical conformational analysis.
J. Pept. Res., 2000, 56, 132-146.
- L. Klaudel, S. Rodziewicz-Motowidło, A. Liwo, K. Rolka.
A comparison of solution conformations of scyliorhinin I and its analogue
with N-methyl-L-phenylalanine in position 7.
Pol. J. Chem., 2000, 74, 1091-1099.
- J. Malicka, M. Groth, J. Karolczak, C. Czaplewski, A. Liwo,
W. Wiczk,
Influence of solvents and leucine configuration at position 5 on
tryptophan fluorescence in cyclic enkephalin analogues.
Biopolymers, 2001, 58, 447-457.
- J. Pillardy, C. Czaplewski, A. Liwo, J. Lee, D.R. Ripoll, R. Kaźmierkiewicz,
St. Ołdziej, W.J. Wedemeyer, K.D. Gibson, Y.A. Arnautova, J. Saunders,
Y.-J. Ye, H.A. Scheraga.
Recent improvements in prediction of protein structure by global
optimization of a potential energy function.
Proc. Natl. Acad. Sci. USA, 2001, 98, 2329-2333.
- J. Malicka, R. Ganzynkowicz, M. Groth, C. Czaplewski, J. Karolczak, A. Liwo,
W. Wiczk.
Fluorescence decay time distribution analysis of cyclic enkephalin
analogues; Influence of solvent and Leu configuration in position 5 on
conformation.
Acta Biochim. Pol., 2001, 48, 95-102.
- M. Groth, J. Malicka, S. Rodziewicz-Motowidło, C. Czaplewski, L. Klaudel,
W. Wiczk, A. Liwo.
Determination of conformational equilibrium of peptides in solution
by NMR spectroscopy and theoretical conformational analysis: application
to the calibration of mean-field solvation models.
Biopolymers, 2001, 60, 79-95.
- A. Liwo, C. Czaplewski, J. Pillardy, H.A. Scheraga,
Cumulant-based expressions for the multibody terms for the correlation between
local and electrostatic interactions in the united-residue force field.
J. Chem. Phys., 2001, 115, 2323-2347.
- J. Pillardy, C. Czaplewski, A. Liwo, W.J. Wedemeyer, J. Lee, D.R. Ripoll,
P. Arlukowicz, S. Ołdziej, Y.A. Arnautova, H.A. Scheraga.
Development of physics-based energy functions that predict medium-resolution
structures for proteins of the
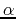 ,
,  and
and 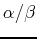 structural
classes. J. Phys. Chem. B, 2001, 7299-7311.
structural
classes. J. Phys. Chem. B, 2001, 7299-7311.
- S. Rodziewicz-Motowidło, A. Lesner, A. Łęgowska, C. Czaplewski, A. Liwo,
K. Rolka, R. Patacchini, L. Quartara.
Synthesis, activity on NK-(3) tachykinin receptor and conformational solution
studies of scyliorhinin II analogs modified at position 16.
J. Pept. Res., 2001, 58, 159-172.
- P. Drabik, A. Liwo, C. Czaplewski, J. Ciarkowski.
The investigation of the effects of counterions in protein dynamics.
Protein Engineering, 2001, 14, 747-752.
- J. Malicka, M. Groth, C. Czaplewski, J. Karolczak, A. Liwo, W. Wiczk.
Influence of solvent and configuration of residues at position 2 and 3 on
distance and mobility of pharmacophore group at position 1 and 4 in cyclic
enkephalin analogues.
Biopolymers, 2001, 59, 180-190.
- C. Czaplewski, D.R. Ripoll, A. Liwo, S. Rodziewicz-Motowidło, R.J. Wawak,
H.A. Scheraga.
Can cooperativity in hydrophobic association be reproduced correctly by
implicit solvation models?
Int. J. Quant. Chem., 2002, 88, 41-55.
- J. Malicka, M. Groth, C. Czaplewski, W. Wiczk, A. Liwo.
Conformational studies of cyclic enkephalin analogues with L- or D-proline
in position 3.
Biopolymers, 2002, 63, 217-231.
- C. Czaplewski, S. Rodziewicz-Motowidło, A. Liwo, D.R. Ripoll, R.J. Wawak,
H.A. Scheraga.
Comment on Ąnti-cooperativity in hydrophobic interactions: A simulation
study of spatial dependence of three-body effects and beyond" [J. Chem.
Phys. 115, 1414 (2001)].
J. Chem. Phys., 2002, 116, 2665-2667.
- A. Liwo, P. Arłukowicz, C. Czaplewski, S. Ołdziej, J. Pillardy,
H.A. Scheraga.
A method for optimizing potential-energy functions by a hierarchical design
of the potential-energy landscape: Application to the UNRES force field.
Proc. Natl. Acad. Sci. U.S.A., 2002, 99, 1937-1942.
- R. Kaźmierkiewicz, A. Liwo, H.A. Scheraga.
Energy-based reconstruction of a protein backbone from its
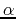 -carbon
trace by a Monte Carlo method.
J. Comput. Chem., 2002, 23, 715-723.
-carbon
trace by a Monte Carlo method.
J. Comput. Chem., 2002, 23, 715-723.
- S. Rodziewicz-Motowidło, K. Brzozowski, A. Łęgowska, A. Liwo, J. Silbering,
M. Smoluch, K. Rolka.
Conformational solution studies of neuropeptide gamma using CD and NMR
spectroscopy.
J. Pept. Sci. 2002, 8, 211-226.
- K. Brzozowski, A. Łęgowska, S. Rodziewicz-Motowidło, A. Liwo, K. Rolka.
The study of conformational equilibrium of c[Gln-Trp-Phe-Gly-Leu-Met], a
NK-2 tachykinin antagonist.
Pol. J. Chem. 2002, 76, 807-814.
- M. Bobrowski, A. Liwo, S. Ołdziej, D. Jeziorek, T. Ossowski.
Ab initio study of the mechanism of singlet-dioxygen addition to
hydroxyaromatic compounds: Negative evidence for the involvement of peroxa
and endoperoxide intermediates.
J. Comput. Chem., 2002, 23, 1076-1089.
- M.O.F. Goulart, P. Falkowski, T. Ossowski, A. Liwo.
Electrochemical study of oxygen interaction with lapachol and its radical
anions. Bioelectrochemistry, 2003, 59, 85-87.
- R. Kaźmierkiewicz, A. Liwo, H.A. Scheraga.
Addition of side chains to a known backbone with defined side-chain centroids.
Biophys. Chem., 2003, 100, 261-280.
- I. Liepiņa, C. Czaplewski, P. Janmey, A. Liwo.
Molecular dynamics study of a gelsolin-derived peptide binding to a lipid
bilayer containing phosphatidylinositol 4,5-bisphospate.
Biopolymers, 2003, 71, 49-70.
- I. Liepiņa, P.A. Janmey, C. Czaplewski, A. Liwo. Molecular dynamics study
of the influence of calcium ions on the conformation of gelsolin S2 domain.
J. Mol. Struct. THEOCHEM, 2003, 630, 309-313.
- C. Czaplewski, S. Rodziewicz-Motowidło, M. Dąbal, A. Liwo, D.R. Ripoll,
H.A. Scheraga.
Molecular simulation study of cooperativity in hydrophobic association:
clusters of four hydrophobic particles.
Biophys. Chem., 2003, 105, 339-359.
- S. Ołdziej, U. Kozłowska, A. Liwo, H.A. Scheraga.
Determination of the potentials of mean force for rotation
about C
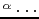 C
C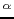 virtual bonds in polypeptides from
the ab initio energy
surfaces of terminally-blocked glycine, alanine, and proline.
J. Phys. Chem. A, 2003, 107, 8035-8046.
virtual bonds in polypeptides from
the ab initio energy
surfaces of terminally-blocked glycine, alanine, and proline.
J. Phys. Chem. A, 2003, 107, 8035-8046.
- K. Maksimiak, S. Rodziewicz-Motowidło, C. Czaplewski, A. Liwo,
H.A. Scheraga.
Molecular simulation study of the potentials of mean force for the
interactions between models of like-charged as well as between charged and
nonpolar amino-acid side chains in water.
J. Phys. Chem. B, 2003, 107, 13496-13504.
- C. Czaplewski, A. Liwo, J. Pillardy, S. Ołdziej, H.A. Scheraga.
Improved Conformational Space Annealing method to treat
 -structure with the UNRES force-field and to enhance scalability
of parallel implementation.
Polymer, 2004, 45, 677-686.
-structure with the UNRES force-field and to enhance scalability
of parallel implementation.
Polymer, 2004, 45, 677-686.
- A. Liwo, S. Ołdziej, C. Czaplewski, U. Kozłowska, H.A. Scheraga.
Parameterization of backbone-electrostatic and multibody contributions
to the UNRES force field for protein-structure prediction from ab initio
energy surfaces of model systems.
J. Phys. Chem. B, 2004, 108, 9421-9438.
- C. Czaplewski, S. Ołdziej, A. Liwo, H.A. Scheraga.
Prediction of the structures of proteins with the UNRES force field,
including dynamic formation and breaking of
disulfide bonds.
Protein Eng. Des. Select., 2004, 17, 29-36.
- K. Guzow, R. Ganzynkowicz, A. Rzeska, J. Mrozek, M. Szabelski, J. Karolczak,
A. Liwo, W. Wiczk.
Photophysical properties of tyrosine and its simple derivatives
studied by time-resolved fluorescence spectroscopy, global analysis
and theoretical calculations.
J. Phys. Chem. B, 2004, 108, 3879-3889.
- M.O.F. Goulart, N.M.F. Lima, A. Euzebio, G. Sant'Ana, P.A.L. Ferraz,
J.C.M. Cavalcanti, P. Falkowski, T. Ossowski, A. Liwo.
Electrochemical studies of isolapachol with emphasis on oxygen interaction with its radical anions.
J. Electroanal. Chem., 2004, 566, 25-29.
- J. Makowska, M. Makowski, A. Giełdoń, A. Liwo, L. Chmurzyński.
Theoretical calculations of heteroconjugation equilibrium constants in systems
moldeling acid-base interactions in side chains of biomolecules using the
potential of mean force.
J. Phys. Chem. B, 2004, 108, 12222-12230.
- M. Khalili, J.A. Saunders, A. Liwo, S. Ołdziej, H.A. Scheraga.
A united-residue force field for calcium-protein interactions.
Prot. Sci., 2004, 13, 2725-2735.
- A. Liwo, P. Arłukowicz, S. Ołdziej, C. Czaplewski,
M. Makowski, H.A. Scheraga.
Optimization of the UNRES force field by hierarchical design of the
potential-energy landscape. 1. Tests of the approach using simple
lattice protein models.
J. Phys. Chem. B., 2004, 108, 16918-16933.
- S. Ołdziej, A. Liwo, C. Czaplewski, J. Pillardy, H.A. Scheraga.
Optimization of the UNRES force field by hierarchical design of the
potential-energy landscape. 2. Off-lattice tests of the method with single
proteins.
J. Phys. Chem. B., 2004, 108, 16934-16949.
- S. Ołdziej, J. Łągiewka, A. Liwo, C. Czaplewski, M. Chinchio,
M. Nanias, H.A. Scheraga.
Optimization of the UNRES force field by hierarchical design of the
potential-energy landscape. 3. Use of many proteins in optimization.
J. Phys. Chem. B., 2004, 108, 16950-16959.
- I. Liepiņa, P. Janmey, C. Czaplewski, A. Liwo.
Towards gelsolin amyloid formation.
Biopolymers, 2004, 76, 543-548.
- J. Makowska, K. Bagińska, F. Kasprzykowski, J.A. Vila, A. Jagielska,
A. Liwo, L. Chmurzyński, H.A. Scheraga.
Interplay of charge distribution and conformation for peptides:
comparison of theory and experiment.
Biopolymers, 2005, 80, 214-224.
- J. Makowska, M. Makowski, A. Liwo, L. Chmurzyński,
Theoretical calculations of homoconjugation equilibrium constants in systems
modeling acid base interactions in side chains of biomolecules using the potential
of mean force.
J. Comput. Chem., 2005, 235-242.
- A. Liwo, M. Khalili, H.A. Scheraga.
Ab initio simulations of protein-folding pathways by molecular dynamics with
the united-residue model of polypeptide chains.
Proc. Natl. Acad. Sci. U.S.A., 2005, 102, 2362-2367.
- C. Czaplewski, A. Liwo, D.R. Ripoll, H.A. Scheraga.
Molecular origin of anticooperativity in hydrophobic association.
J. Phys. Chem. B, 2005, 109, 8108-8119.
- S. Ołdziej, C. Czaplewski, A. Liwo, M. Chinchio, M. Nanias, J.A.
Vila, M. Khalili, Y. A. Arnautova, A. Jagielska, M. Makowski, H. D.
Schafroth, R. Kaźmierkiewicz, D. R. Ripoll, J. Pillardy, J.A.
Saunders, Y.K. Kang, K.D. Gibson, H.A. Scheraga.
Physics-based protein-structure prediction using a hierarchical protocol
based on the UNRES force field: Assessment in two blind tests.
Proc. Natl. Acad. Sci. U.S.A., 2005, 102, 7547-7552.
- M. Khalili, A. Liwo, F. Rakowski, P. Grochowski, H.A. Scheraga.
Molecular dynamics with the united-residue model of polypeptide chains.
I. Lagrange equations of motion and tests of numerical stability in the
microcanonical mode, J. Phys. Chem. B, 2005, 109, 13785-13797.
- M. Khalili, A. Liwo, A. Jagielska, H.A. Scheraga.
Molecular dynamics with the united-residue model of polypeptide chains.
II. Langevin and Berendsen-bath dynamics and tests on model
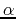 -helical
systems. J. Phys. Chem. B, 2005, 109, 13798-13810.
-helical
systems. J. Phys. Chem. B, 2005, 109, 13798-13810.
- I. Liepina, P.A. Janmey, C. Czaplewski, A. Liwo.
Membrane initiated gelsolin amyloid formation.
Biopolymers, 2005, 80, 596-597.
- C. Czaplewski, S. Kalinowski, A. Liwo, H. A. Scheraga.
Comparison of two approaches to potential of mean force calculations of
hydrophobic association: particle insertion and weighted histogram analysis
methods.
Mol. Phys., 2005, 103, 3153-3167.
- C. Czaplewski, S. Kalinowski, A. Liwo, D.R. Ripoll, H.A. Scheraga.
Reply to ``Comment on 'Molecular Origin of Anticooperativity in
Hydrophobic Association' ''
J. Phys. Chem. B., 2005, 109, 21222-21224.
- M. Khalili, A. Liwo, H.A. Scheraga.
Kinetic studies of folding of the B-domain of staphylococcal protein A
with molecular dynamics and a united-residue (UNRES) model of polypeptide
chains.
J. Mol. Biol., 2006, 355, 536-547.
- J. Makowska, S. Rodziewicz-Motowidło, K. Bagińska, J.A. Vila, A. Liwo,
L. Chmurzyński, H.A. Scheraga.
Polyproline II conformation is one of many local conformational states and
is not an overall conformation of unfolded peptides and proteins.
Proc. Natl. Acad. Sci. USA, 2006, 103, 1744-1749.
- J. Makowska, K. Baginńska, M. Makowski, A. Jagielska, A. Liwo,
F. Kasprzykowski, L. Chmurzynński, H.A. Scheraga.
Assessment of two theoretical methods to estimate potentiometric titration
curves of peptides: comparison with experiment.
J. Phys. Chem. B, 2006, 110, 4451-4458.
- I. Liepina, S. Ventura, C. Czaplewski, A. Liwo.
Molecular dynamics study of amyloid formation of two Abl-SH3 domain peptides.
J. Pept. Sci., 2006, 12, 780-789.
- F. Rakowski, P. Grochowski, B. Lesyng, A. Liwo, H. A. Scheraga.
Implementation of a symplectic multiple-time-step molecular dynamics algorithm,
based on the united-residue mesoscopic potential energy function.
J. Chem. Phys., 2006, 125, 204107.
- A. Liwo, M. Khalili, C. Czaplewski, S. Kalinowski, S. Ołdziej, K. Wachucik,
H.A. Scheraga.
Modification and optimization of the united-residue (UNRES) potential energy
function for canonical simulations. I. Temperature dependence of the effective
energy function and tests of the optimization method with single training
proteins.
J. Phys. Chem. B, 2007, 111, 260-285.
- A.V. Rojas, A. Liwo, H.A. Scheraga.
Molecular dynamics with the united-residue force field: Ab Initio folding
simulations of multichain proteins.
J. Phys. Chem. B., 2007, 111, 293-309.
- M. Makowski,A. Liwo, H.A. Scheraga.
Simple physics-based analytical formulas for the potentials of mean force for
the interaction of amino acid side chains in water. 1. Approximate expression
for the free energy of hydrophobic association based on a Gaussian-overlap
model. J. Phys. Chem. B., 2007, 111, 2910-2916.
- M. Makowski, A. Liwo, K. Maksimiak, J. Makowska, H.A. Scheraga.
Simple physics-based analytical formulas for the potentials of mean force for
the interaction of amino acid side chains in water. 2. Tests with simple
spherical systems.
J. Phys. Chem. B., 2007, 111, 2917-2924.
- M. Makowski, E. Sobolewski, C. Czaplewski, A. Liwo, S. Ołdziej, J.H. No,
H.A. Scheraga.
Simple physics-based analytical formulas for the potentials of mean force for
the interaction of amino acid side chains in water. 3. Calculation and
parameterization of the potentials of mean force of pairs of identical
hydrophobic side chains.
J. Phys. Chem. B., 2007, 111, 2925-2931.
- M. Bobrowski, A. Liwo, K. Hirao.
Theoretical study of the energetics of the reactions of triplet dioxygen
with hydroquinone, semiquinone, and their protonated forms: relation to
the mechanism of superoxide generation in the respiratory chain.
J. Phys. Chem. B., 2007, 111, 3543-3549.
- J. Makowska, S. Rodziewicz-Motowidło, K. Bagińska, M. Makowski, J.A. Vila,
A. Liwo, L. Chmurzyński, H.A. Scheraga.
Further evidence for the absence of polyproline II stretch in the XAO peptide.
Biophys. J., 2007, 92, 2904-2917.
- H. Grajek, A. Liwo, W. Wiczk, G. Żurkowska.
Resolution of the excitation-emission spectra of FMN in rigid
poly(vinyl alcohol) matrices.
J. Photochem. Photobiol. B, 2007, 86, 193-198.
- M.Y. Niv, D.R. Ripoll, J.A. Vila, A. Liwo, S. Vanamee, A.K.
Aggarwal, H. Weinstein, H.A. Scheraga.
Topology of Type II REases revisited; structural classes and
the common conserved core.
Nucl. Acids Res., 2007, 35, 2227-2237.
- U. Kozłowska, A. Liwo, H.A. Scheraga.
Determination of virtual-bond-angle potentials of mean force for coarse-grained
simulations of protein structure and folding from ab initio energy surfaces of
terminally-blocked glycine, alanine, and proline.
J. Phys.: Condens. Matter, 2007, 19, 285203.
- M. Chinchio, C. Czaplewski, A. Liwo, S. Ołdziej, H.A. Scheraga.
Dynamic formation and breaking of disulfide bonds in molecular dynamics
simulations with the UNRES force field.
J. Chem. Theory and Comput., 2007, 3, 1236-1248.
- E. Sobolewski, M. Makowski, C. Czaplewski, A. Liwo, S. Ołdziej, H.A. Scheraga.
Potential of mean force of hydrophobic association: Dependence on solute size.
J. Phys. Chem. B, 2007, 111, 10765-10774.
- R.K. Murarka, A. Liwo, H.A. Scheraga.
Separation of time scale and coupling in the motion governed by the coarse-grained
and fine degrees of freedom in a polypeptide backbone.
J. Chem. Phys., 2007, 127, 155103.
- T. Ossowski, M.O.F. Goulart, F.C. de Abreu, A.E.G. Sant'Ana, D. Zarzeczańska,
P.R.B. Miranda, C.D. Costa, A. Liwo, P. Falkowski. Determination of the pK
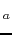 values of some biologically active and inactive hydroxyquinones.
J. Braz. Chem. Soc., 2008, 19, 175-183.
values of some biologically active and inactive hydroxyquinones.
J. Braz. Chem. Soc., 2008, 19, 175-183.
- D.S. Kleinerman, C. Czaplewski, A. Liwo, H.A. Scheraga.
Implementations of Nosé-Hoover and Nosé-Poincaré termostats in mesoscopic
dynamic simulations with the united-residue model of a polypeptide chain.
J. Chem. Phys., 2008, 128, 245103.
- J. Makowska, K. Bagińska, A. Liwo, L. Chmurzyński, H.A. Scheraga.
Acidic-basic properties of three alanine-based peptides containing
acidic and basic side chains: Comparison between theory and experiment.
Biopolymers: Peptide Science, 2008, 90, 724-732.
- J. Makowska, K. Bagińska, A. Skwierawska, A. Liwo, L. Chmurzyński,
H.A. Scheraga.
Influence of charge and size of terminal amino-acid residues on local
conformational states and shape of alanine-based peptides.
Biopolymers: Peptide Science, 2008, 90, 724-732.
- M. Makowski, E. Sobolewski, C. Czaplewski, S. Ołdziej, A. Liwo,
H.A. Scheraga.
Simple physics-based analytical formulas for the potentials of mean force
for the interaction of amino acid side chains in water. IV. Pairs of
different hydrophobic side chains.
J. Phys. Chem. B, 2008, 112, 11385-11395.
- H. Shen, C. Czaplewski, A. Liwo, H.A. Scheraga.
Implementation of a Serial Replica Exchange Method in a Physics-Based
United-Residue (UNRES) Force Field.
J. Chem. Theory and Comput., 2008, 4, 1386-1400.
- A. Skwierawska, S. Rodziewicz-Motowidło, S. Ołdziej, A. Liwo,
H.A. Scheraga.
Conformational studies of the
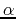 -helical 28-43 fragment of the B3
domain of the immunoglobulin binding protein G from Streptococcus.
Biopolymers: Peptide Science, 2009, 91, 37-51.
-helical 28-43 fragment of the B3
domain of the immunoglobulin binding protein G from Streptococcus.
Biopolymers: Peptide Science, 2009, 91, 37-51.
- S. Rodziewicz-Motowidło, J. Iwaszkiewicz, R. Sosnowska, P. Czaplewska,
E. Sobolewski, A. Szymańska, K. Stachowiak, A. Liwo.
The role of the Val57 amino-acid residue in the hinge loop of the human
cystatin C. Conformational studies of the
 2-L1-
2-L1- 3 segments of wild
type human cystatin C and its mutants.
Biopolymers, 2009, 91, 373-383.
3 segments of wild
type human cystatin C and its mutants.
Biopolymers, 2009, 91, 373-383.
- C. Czaplewski, S. Kalinowski, A. Liwo, H.A. Scheraga.
Application of multiplexed replica exchange molecular dynamics
to the UNRES force field: tests with
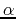 and
and 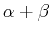 proteins.
J. Chem. Theor. Comput., 2009, 5, 627-640.
proteins.
J. Chem. Theor. Comput., 2009, 5, 627-640.
- G.G. Maisuradze, A. Liwo, H.A. Scheraga.
Principal component analysis for protein folding dynamics.
J. Mol. Biol., 2009, 385, 312-329.
- A. Skwierawska, J. Makowska, S. Ołdziej, A. Liwo, H.A. Scheraga.
Mechanism of formation of the C-terminal
 -hairpin of the B3 domain
of the immunoglobulin binding protein G from Streptococcus.
I. Importance of hydrophobic interactions in stabilization of
-hairpin of the B3 domain
of the immunoglobulin binding protein G from Streptococcus.
I. Importance of hydrophobic interactions in stabilization of  -hairpin
structure.
Proteins: Strut. Funct. Bioinfo., 2009, 75, 931-953.
-hairpin
structure.
Proteins: Strut. Funct. Bioinfo., 2009, 75, 931-953.
- A. Skwierawska, W. Żmudzińska, S. Ołdziej, A. Liwo, H.A. Scheraga.
Mechanism of formation of the C-terminal
 -hairpin of the B3 domain
of the immunoglobulin binding protein G from Streptococcus. II.
Interplay of local backbone conformational dynamics and long-range
hydrophobic interactions in hairpin formation.
Proteins: Strut. Funct. Bioinfo., 2009, 76, 637-654.
-hairpin of the B3 domain
of the immunoglobulin binding protein G from Streptococcus. II.
Interplay of local backbone conformational dynamics and long-range
hydrophobic interactions in hairpin formation.
Proteins: Strut. Funct. Bioinfo., 2009, 76, 637-654.
- H. Shen, A. Liwo, H.A. Scherega.
An improved functional form for the temperature scaling
factors of the components of the mesoscopic UNRES force field for
simulations of protein structure and dynamics.
J. Phys. Chem. B, 2009, 113, 8738-8744.
- G.G. Maisuradze, A. Liwo, H.A. Scheraga.
How adequate are one- and two-dimensional free energy landscapes for
protein folding dynamics?
Phys. Rev. Lett., 2009, 102, 238102.
- Y. He, Y. Xiao, A. Liwo, H.A. Scheraga.
Exploring the parameter space of the coarse-grained
UNRES force field by random search: selecting a transferable medium-resolution
force field.
J. Comput. Chem., 2009, 30, 2127-2135.
- E. Sobolewski, M. Makowski, S. Ołdziej, C. Czaplewski, A. Liwo,
H.A. Scheraga.
Towards temperature-dependent coarse-grained potentials of side-chain
interactions for protein folding simulations. I. Molecular dynamics
study of a pair of methane molecules in water at various temperatures.
PEDS, 2009, 22, 547-552.
- A. Lewandowska, S. Ołdziej, A. Liwo, H.A. Scheraga.
Mechansm of formation of the C-terminal
 -hairpin of the B3 domain of the immunoglobulin binding protein G from
Streptococcus. Part III. Dynamics of long-range hydrophobic interactions.
Proteins: Struct. Funct. and Bioinf., 2010, 78, 723-737.
-hairpin of the B3 domain of the immunoglobulin binding protein G from
Streptococcus. Part III. Dynamics of long-range hydrophobic interactions.
Proteins: Struct. Funct. and Bioinf., 2010, 78, 723-737.
- M. Makowski, C. Czaplewski, A. Liwo, H.A. Scheraga.
Potential of mean force of association of large hydrophobic particles: toward the
nanoscale limit. J. Phys. Chem. B, 2010, 114, 993-1003.
- G. Maisuradze, A. Liwo, H.A. Scheraga.
Relation between free energy landscapes of proteins and dynamics.
J. Chem. Theor. Comput., 2010, 6, 583-595.
- A. Liwo, S. Ołdziej, C. Czaplewski, D.S. Kleinerman, P. Blood,
H.A. Scheraga.
Implementation of molecular dynamics and its extensions
with the coarse-grained UNRES force field on massively
parallel systems; towards millisecond-scale simulations of
protein structure, dynamics, and thermodynamics
J. Chem. Theor. Comput., 2010, 6, 890-909.
- U. Kozłowska, A. Liwo. H.A. Scheraga.
Determination of side-chain-rotamer and side-chain and backbone
virtual-bond-stretching potentials of mean force from AM1 energy surfaces of
terminally-blocked amino-acid residues, for coarse-grained simulations of
protein structure and folding. 1. The Method.
J. Comput. Chem., 2010, 31, 1143-1153.
- U. Kozłowska, G.G. Maisuradze, A. Liwo, H.A. Scheraga.
Determination of side-chain-rotamer and side-chain and backbone
virtual-bond-stretching potentials of mean force from AM1 energy surfaces of
terminally-blocked amino-acid residues, for coarse-grained simulations of
protein structure and folding. 2. Results, comparison with statistical
potentials, and implementation in the UNRES force field.
J. Comput. Chem., 2010, 31, 1154-1167.
- A. Lewandowska, S. Ołdziej, A. Liwo, H.A. Scheraga.
Mechanism of formation of the C-terminal
 -hairpin of the B3 domain of
the immunoglobulin binding protein G from Streptococcus. Part IV.
Implication for the mechanism of folding of the parent protein.
Biopolymers, 2010, 93, 469-480.
-hairpin of the B3 domain of
the immunoglobulin binding protein G from Streptococcus. Part IV.
Implication for the mechanism of folding of the parent protein.
Biopolymers, 2010, 93, 469-480.
- G.G. Maisuradze, P. Senet, C. Czaplewski, A. Liwo, H.A. Scheraga.
Investigation of protein folding by coarse-grained molecular dynamics with
the UNRES force field. J. Phys. Chem. A, 2010, 114, 4471-4485.
- M. Maciejczyk, A. Liwo, H.A. Scheraga.
Coarse-grained model of nucleic acid bases.
J. Comput. Chem., 2010, 31, 1644-1655.
- G.G. Maisuradze, A.Liwo, S. Ołdziej, H.A. Scheraga.
Evidence, from simulations, of a single state with residual native structure
at the thermal denaturation midpoint of a small globular protein.
J. Am. Chem. Soc, 2010, 132, 9444-9452.
- A. Rojas, A. Liwo, D. Browne, H. A. Scheraga,
Mechanism of fiber assembly; treatment of A
 -peptide
peptide aggregation with a coarse-grained united-residue force field.
J. Mol. Biol., 2010, 404, 537-552.
-peptide
peptide aggregation with a coarse-grained united-residue force field.
J. Mol. Biol., 2010, 404, 537-552.
- M. Kozak, A. Lewandowska, S. Ołdziej, S. Rodziewicz-Motowidło, A. Liwo,
Combination of SAXS and NMR techniques as a tool for the determination of
peptide structure in solution.
J. Phys. Chem. Letters, 2010, 1, 3128-3131.
- Y. He, A. Liwo, H. Weinstein, H.A. Scheraga,
PDZ Binding to the BAR Domain of PICK1 is Elucidated by Coarse-grained Molecular Dynamics.
J. Mol. Biol., 2011, 405, 298-314.
- M. Makowski, A. Liwo, E. Sobolewski, H.A. Scheraga.
Simple physics-based analytical formulas for the potentials of mean force of the
interaction of amino-acid side chains in water. V. Like-charged side chains.
J. Phys. Chem. B, 2011, 115, 6119-6129.
- M. Makowski, A. Liwo, H.A. Scheraga.
Simple physics-based analytical formulas for the potentials of mean force of the
interaction of amino-acid side chains in water. VI. Oppositely charged side chains.
J. Phys. Chem. B, 2011, 115, 6130-6137.
- A. Rojas, A. Liwo, H.A. Scheraga.
A study of the
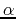 -helical intermediate preceding the aggregation of
the amino-terminal fragment of the A
-helical intermediate preceding the aggregation of
the amino-terminal fragment of the A -amyloid peptide (1-28).
J. Phys. Chem. B, 2011, 115, 12978-12983.
-amyloid peptide (1-28).
J. Phys. Chem. B, 2011, 115, 12978-12983.
- J. Makowska, A. Liwo, W. Żmudzińska, A. Lewandowska, L. Chmurzyński, H.A. Scheraga.
Like-charged residues at the ends of oligoalanine sequences might induce a chain reversal.
Biopolymers, 2012, 97, 240-249.
- Y. Yin, G.G. Maisuradze, A. Liwo, H.A. Scheraga.
Hidden protein folding pathways in free-energy landscapes uncovered by network
analysis.
J. Chem. Theory Comput., 2012, 8, 1176-1189.
- A.K. Sieradzan, H.A. Scheraga, A. Liwo.
Determination of effective potentials for the stretching of C
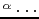 C
C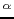 virtual bonds in polypeptide chains for coarse-grained simulations of proteins from ab
initio energy surfaces of N-Methylacetamide and N-Acetylpyrrolidine.
J. Chem. Theor. Comput., 2012, 8, 1334-1343.
virtual bonds in polypeptide chains for coarse-grained simulations of proteins from ab
initio energy surfaces of N-Methylacetamide and N-Acetylpyrrolidine.
J. Chem. Theor. Comput., 2012, 8, 1334-1343.
- E. Golas, G.G. Maisuradze, P. Senet, S. Ołdziej, C. Czaplewski, H. A. Scheraga, A. Liwo.
Simulation of the opening and closing of Hsp70 chaperones by coarse-grained molecular dynamics.
J. Chem. Theor. Comput., 2012, 8, 1750-1764.
- E. Sobolewski, S. Ołdziej, M. Wiśniewska, A. Liwo, M. Makowski.
Toward temperature-dependent coarse-grained potentials of side-chain interactions for
protein folding simulations. II. Molecular dynamics study of pairs of different types of
interactions in water at various temperatures
J. Phys. Chem. B, 2012, 23, 6844-6853.
- A. Hałabis, W. Żmudzińska, A. Liwo, S. Ołdziej.
Conformational dynamics of the Trp-Cage miniprotein at its folding temperature.
J. Phys. Chem. B, 2012, 23, 6898-6907.
- G.G. Maisuradze, R. Zhou, A. Liwo, Y. Xiao, H.A. Scheraga. Effects
of Mutation, Truncation, and Temperature on the Folding Kinetics of a
WW Domain.
J. Mol. Biol., 2012, 420, 350-365.
- A.K. Sieradzan, A. Liwo, U.H.E. Hansmann.
Folding and self-assembly of a small protein complex.
J. Chem. Theory Comput., 2012, 8, 3416-3422.
- A. Krokhotin, A. Liwo, A.J. Niemi, H.A. Scheraga.
Coexistence of phases in a protein heterodimer.
J. Chem. Phys., 2012, 137, 035101.
- A.K. Sieradzan, U.H.E. Hansmann, H.A. Scheraga, A. Liwo.
Extension of UNRES force field to treat polypeptide chains with D-amino-acid residues.
J. Chem. Theory Comput., 2012, 8, 4746-4757.
- D. Lapidus, V. Duka, V. Stonkus, C. Czaplewski, A. Liwo, S. Ventura, I. Liepina.
Multiple b-Sheet Molecular Dynamics of Amyloid Formation From Two Abl-SH3 Domain Peptides.
Biopolymers: Peptide Science, 2012, 86, 557-566.
- M. Makowska, A. Liwo, L. Chmurzyński, H.A. Scheraga,
Influence of the Length of the Alanine Spacer on the Acidic-Basic Properties of
the Ac-Lys-(Ala)
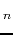 -Lys-NH
-Lys-NH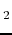 Peptides (n=0, 1, 2, ..., 5),
J. Sol. Chem., 2012, 41, 1738-1746.
Peptides (n=0, 1, 2, ..., 5),
J. Sol. Chem., 2012, 41, 1738-1746.
- Y. He, M. Maciejczyk, S. Ołdziej, H.A. Scheraga, A, Liwo.
Mean-field interactions between nucleic-acid-base
dipoles can drive the formation of a double helix,
Phys. Rev. Lett., 2013, 110, 098101.
- A. Liwo.
Coarse graining: a tool for large-scale simulations or more?
Phys. Scr., 2013, 87, 058502.
- G. Maisuradze, A. Liwo, P. Senet, H.A. Scheraga.
Local vs global motions in protein folding,
J. Chem. Theory Comput., 2013, 9, 2907-2921.
- Y. He, M.A. Mozolewska, P. Krupa, A.K. Sieradzan, T.K. Wirecki, A. Liwo,
K. Kachlishvili, S. Rackovsky, D. Jagieła, R. Ślusarz, C.R. Czaplewski,
S. Ołdziej, H.A. Scheraga,
Lessons from application of the UNRES force field to predictions of structures
of CASP10 targets.
Proc. Natl. Acad. Sci. U.S.A., 2013, 110, 14936-14941.
- P. Krupa, A.K. Sieradzan, S. Rackovsky, M. Baranowski, S. Ołdziej,
H.A. Scheraga, A. Liwo, C. Czaplewski.
Improvement of the Treatment of Loop Structures in the UNRES
Force Field by Inclusion of Coupling between Backbone- and
Side-Chain-Local Conformational States
J. Chem. Theory Comput., 2013, 4620-4632.
- A. Krokhotin, A. Liwo, G.G. Maisuradze, A.J. Niemi, H.A. Scheraga.
Kinks, loops, and protein folding, with protein A as an example.
J. Chem. Phys., 2014, 140, 025101.
- A. K. Sieradzan, A. Niadzvedtski, H.A. Scheraga, A. Liwo,
Revised backbone-virtual-bond-angle potentials to treat the L- and D-amino
acid residues in the coarse-grained united residue (UNRES) force field.
J. Chem. Theory Comput., 2014, 10, 2194-2203.
- K. Kachlishvili, G.G. Maisuradze, O.A. Martin, A. Liwo, J.A. Vila,
H.A. Scheraga.
Accounting for a mirror-image conformation as a subtle effect in protein
folding.
Proc. Natl. Acad. Sci. USA, 2014, 111, 8458-8463.
- A. Liwo, M. Baranowski, C. Czaplewski, E. Gołaś, Y. He, D. Jagieła,
P. Krupa, M. Maciejczyk, M. Makowski, M.A Mozolewska, A. Niadzvedtski,
S. Ołdziej, H.A. Scheraga, A.K. Sieradzan, R. Ślusarz, T. Wirecki,
Y. Yin, B. Zaborowski.
A unified coarse-grained model of biological macromolecules based on
mean-field multipole.multipole interactions.
J. Molec. Model., 2014, 20, 1-15.
- G.A. Khoury, A. Liwo, F. Khatib, H. Zhou, G. Chopra, J. Bacardit,
L.O. Bortot, R.A. Faccioli, X. Deng, Y. He, P. Krupa, J. Li, M.A. Mozolewska,
A.K. Sieradzan, J. Smadbeck, T. Wirecki, S. Cooper, J. Flatten,
K. Xu, D. Baker, J. Cheng, A.C.B. Delbem, C.A. Floudas, C. Keasar,
M. Levitt, Z. Popovič, H.A. Scheraga, J. Skolnick, S.N. Crivelli1, and
Foldit Players.
WeFold: A coopetition for protein structure prediction.
Proteins: Struct. Funct. Bioinfo., 2014, 82, 1850-1868.
- M. Maciejczyk, A. Spasic, A. Liwo, H.A. Scheraga
DNA Duplex Formation with a Coarse-Grained Model.
J. Chem. Theory Comput., 2014, 10, 5020-5035.
- R. Zhou, G.G. Maisuradze, D. Suol, T. Todorovski, M.J. Macias, Y. Xiao,
H.A. Scheraga, C. Czaplewski, A. Liwo.
Folding kinetics of WW domains with the united residue force field for bridging
microscopic motions and experimental measurements.
Proc. Natl. Acad. Sci. U.S.A, 2014, 111, 18243-18248.
- E.I. Gołaś, C. Czaplewski, H.A. Scheraga, and A. Liwo.
Common functionally important motions of the nucleotide-binding domain of
Hsp70.
Proteins: Struct. Func. Bioinf., 2015, 83, 282-299.
- A.G. Lipska, A.K. Sieradzan, P. Krupa, M. Mozolewska, S. D'Auria, A. Liwo.
Studies of conformational changes of an arginine-binding protein
from Thermotoga maritima in the presence and absence of ligand
via molecular dynamics simulations with the coarse-grained
UNRES force field.
J. Molec. Modeling, 2015, 21,10.1007/s00894-015-2609-1
- A.K. Sieradzan, P. Krupa, H.A. Scheraga, A. Liwo, C. Czaplewski.
Physics-Based Potentials for the Coupling between Backbone- and
Side-Chain-Local Conformational States in the United Residue
(UNRES) Force Field for Protein Simulations.
J. Chem. Theory Comput., 2015, 11, 817-831.
- P. Krupa, M.A. Mozolewska, K. Joo, J. Lee, C. Czaplewski, A. Liwo.
Prediction of Protein Structure by Template-Based Modeling Combined
with the UNRES Force Field.
J. Chem. Inf. Model., 2015, 55, 1271-1281.
- Y. Yin, A.K. Sieradzan, A. Liwo, Y. He, H.A. Scheraga.
Physics-Based Potentials for Coarse-Grained Modeling of Protein DNA
Interactions.
J. Chem. Theory Comput., 2015, 11, 1792-1808.
- M.A. Mozolewska, P. Krupa, H.A. Scheraga, A. Liwo.
Molecular modeling of the binding modes of the iron-sulfur protein
to the Jac1 co-chaperone from Saccharomyces cerevisiae by all-atom
and coarse-grained approaches.
Proteins: Struct. Funct. Bioinfo., 2015, 83, 1414-1426.
- Y. He, A. Liwo, H.A. Scheraga.
Optimization of a Nucleic Acids united-RESidue 2-Point model (NARES-2P) with a
maximum-likelihood approach.
J. Chem. Phys., 2015, 143, 243111.
- B. Zaborowski, D. Jagieła, C. Czaplewski, A. Hałabis, A. Lewandowska, W. Żmudzińska,
S. Ołdziej, A. Karczyńska, C. Omieczynski, T. Wirecki, A. Liwo.
A maximum-likelihood approach to force-field calibration.
J. Chem. Inf. Model., 2015, 55, 2050-2070.
- P. Krupa, M.A. Mozolewska, M. Wiśniewska, Y. Yin, Y. He, A.K. Sieradzan, R. Ganzynkowicz, A.G. Lipska, A. Karczyńska,
M. Ślusarz, R. Ślusarz, A. Giełdoń, C. Czaplewski, D. Jagieła, B. Zaborowski, H.A. Scheraga, A. Liwo.
Performance of protein-structure predictions with the physics-based UNRES force field in CASP11.
Bioinformatics, 2016, 32, 3270-3278.
- A.G. Lipska, S.R. Seidman, A.K. Sieradzan, A. Giełdoń, A. Liwo, H.A. Scheraga.
Molecular dynamics of protein A and a WW domain with a united-residue model including hydrodynamic interaction.
J. Chem. Phys., 2016, 144, 184110.
- M.A. Mozolewska, P. Krupa, B. Zaborowski, A. Liwo, J. Lee, K. Joo, C. Czaplewski.
Use of Restraints from Consensus Fragments of Multiple Server Models to Enhance Protein-Structure
Prediction Capability of the UNRES Force Field.
J. Chem. Inf. Model., 2016, 56, 2263-2279.
- M.A. Mozolewska, A.K. Sieradzan, A. Niadzvedstki, C. Czaplewski, A. Liwo, P. Krupa.
Role of the sulfur to
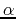 -carbon thioether bridges in thurincin H.
J. Biomol. Struct. Dyn., 2017, 35, 2868-2879.
-carbon thioether bridges in thurincin H.
J. Biomol. Struct. Dyn., 2017, 35, 2868-2879.
- M. Makowski, A. Liwo, H.A. Scheraga.
Simple physics-based analytical formulas for the potentials of mean force of the interaction of
amino acid side chains in water. VII. Charged-hydrophobic/polar and polar-hydrophobic/polar side
chains.
J. Phys. Chem. B, 2017, 121, 379-390.
- T. Johnston, B. Zhang, A. Liwo, S. Crivelli, M. Taufer.
In situ data analytics and indexing of protein trajectories.
J. Comput. Chem., 2017, 38, 1419-1430.
- A.K. Sieradzan, M. Makowski, A. Augustynowicz, A. Liwo.
A general method for the derivation of the functional forms of the effective energy
terms in coarse-grained energy functions of polymers. I. Backbone potentials of
coarse-grained polypeptide chains.
J. Chem. Phys., 2017, 146, 124106.
- P. Krupa, A.K. Sieradzan, M.A. Mozolewska, H. Li, A. Liwo, H.A. Scheraga.
Dynamics of Disulfide-Bond Disruption and Formation in the Thermal Unfolding of Ribonuclease A.
J. Chem. Theory Comput., 2017, 13, 5721-5730.
- P. Krupa, A. Hałabis, W. Żmudzińska, S. Ołdziej, H.A. Scheraga, A. Liwo.
Maximum Likelihood Calibration of the UNRES Force Field for
Simulation of Protein Structure and Dynamics.
J. Chem. Inf. Model. 2017, 57, 2364-2377
- E.A. Lubecka, A. Liwo.
A general method for the derivation of the functional forms of the effective
energy terms in coarse-grained energy functions of polymers.
II. Backbone-local potentials of coarse-grained O1
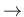 4-bonded
polyglucose chains.
J. Chem. Phys. 2017, 146, 124106.
4-bonded
polyglucose chains.
J. Chem. Phys. 2017, 146, 124106.
- A.S. Karczyńska, C. Czaplewski, P. Krupa, M.A. Mozolewska, K. Joo, J. Lee, A. Liwo.
Ergodicity and Model Quality in Template-Restrained Canonical and Temperature/Hamiltonian
Replica Exchange Coarse-Grained Molecular Dynamics Simulations of Proteins
J. Comput. Chem.,2017, 38, 2730-2746.
- A.S. Karczyńska, M.A. Mozolewska, P. Krupa, A. Giełdoń, A. Liwo, C. Czaplewski.
Prediction of protein structure with the coarse-grained UNRES
force field assisted by small X-ray scattering data and
knowledge-based information.
Proteins: Struct. Func. Bioinfo., 2018, 86 (S1), 228-239.
- M.J. Grzeszczuk, A. Bąk, A.M. Banaś, P. Urbanowicz, S. Dunin-Horkawicz,
. Gieldon, C. Czaplewski, A. Liwo, E.K. Jagusztyn-Krynicka.
Impact of selected amino acids of HP0377 (Helicobacter pylori thiol oxidoreductase)
on its functioning as a CcmG (cytochrome c maturation) protein and Dsb (disulfide bond) isomerase.
PLoS ONE, 2018, 13(4): e0195358.
- C. Czaplewski, A. Karczyńska, A.K. Sieradzan, A. Liwo.
UNRES server for physics-based coarse-grained simulations and prediction
of protein structure, dynamics and thermodynamics.
Nucleic Acids Research, 2018, 46, W304-W309.
- A. Karczyńska, M.A. Mozolewska, P. Krupa, A. Giełdoń, K.K. Bojarski, B. Zaborowski,
A. Liwo, R. Ślusarz, M. Ślusarz, J. Lee, K. Joo, Cezary Czaplewski.
Use of the UNRES force field in template-based prediction of protein structures
and the refinement of server models: Test with CASP12 targets.
J. Molec. Graphics Modeling, 2018, 83, 92-99.
- C. Keasar, L.J. McGuffin, E.B. Wallner, G. Chopra, B. Adhikari, D. Bhattacharya, L. Blake, L.O. Bortot, R.Z. Cao, B.K. Dhanasekaran,
I. Dimas, R.A. Faccioli, E. Faraggi, R. Ganzynkowicz, S. Ghosh, S. Ghosh, A. Giełdoń, Ł. Golon, Y. He, L. Heo, J. Hou, J M. Khan,
F. Khatib, G.A. Khoury, C. Kieslich, D.E. Kim, P. Krupa, G.R. Lee, H.B. Li, J.L. Li, A. Lipska, A. Liwo, A.H.A. Maghrabi, A. Hassan,
M. Mirdita, S. Mirzaei, M.A. Mozolewska, M. Onel, S. Ovchinnikov, A. Shah, U. Shah, T. Sidi, A.K. Sieradzan, M. Ślusarz, R. Ślusarz,
J. Smadbeck, P. Tamamis, T.N. Phanourios, T. Wirecki, Y.P. Yin, Y. Zhang, J. Bacardit, M. Baranowski, N. Chapman, S. Cooper,
A. Defelicibus, J. Flatten, B. Koepnick, Z. Popovic, B. Zaborowski, D. Baker, J.L. Cheng, C. Czaplewski, A.C.B. Delbem,
A. Claudio, C. Floudas, A. Kloczkowski, S. Ołdziej, M. Levitt, H.A. Scheraga, C. Seok, J. Söding, S. Vishveshwara, D. Xu, S.N. Crivelli,
Foldit Players.
An analysis and evaluation of the WeFold collaborative for protein structure prediction and its pipelines in CASP11 and CASP12.
Sci. Rep., 2018, 8, 9939.
- S.D. Vallet, A.E. Miele, U. Uciechowska-Kaczmarzyk, A. Liwo, B. Duclos,
S.A. Samsonov, S. Ricard-Blum.
Insights into the structure and dynamics of lysyl oxidase propeptide,
a flexible protein with numerous partners.
Sci. Rep, 2018, 8, 11768.
- A.K. Sieradzan, Ł. Golon, A. Liwo.
Prediction of DNA and RNA structure with the NARES-2P force field and conformational
space annealing.
Phys. Chem. Chem. Phys., 2018, 20, 19656-19663.
- G. Kohut, A. Liwo, S. Bősze, T. Beke-Somfai, S.A. Samsonov.
Protein-Ligand Interaction Energy-Based Entropy Calculations: Fundamental Challenges For Flexible Systems.
J. Phys. Chem. B, 2018, 122, 7821-7827.
- A.K. Sieradzan, A. Giełdoń, Y. Yin, Y. He, H.A. Scheraga, A. Liwo.
A new protein nucleic-acid coarse-grained force field based on the UNRES and NARES-2P force fields.
J. Comput. Chem., 2018, 29, 2345-2349.
- E. Faraggi, P. Krupa, M.A. Mozolewska, A. Liwo, A. Kloczkowski.
Reoptimized UNRES potential for protein model quality assessment.
Genes, 2018, 9, 601.
- A. Liwo, A. K. Sieradzan, A. G. Lipska, C. Czaplewski, I. Joung,
W. Żmudzińska, A. Hałabis, S. Ołdziej.
A general method for the derivation of the functional forms of the effective
energy terms in coarse-grained energy functions of polymers.
III. Determination of scale-consistent backbone-local and correlation
potentials in the UNRES force field and force-field calibration and validation.
J. Chem. Phys., 2019, 150, 155104.
- S. A. Samsonov, E. A. Lubecka, K. K. Bojarski, R. Ganzynkowicz,
A. Liwo.
Local and long range potentials for heparin-protein systems for
coarse-grained simulations.
Biopolymers. 2019, e23269, https://doi.org/10.1002/bip.23269
(Early View).
- E. Lubecka, A. Liwo.
Use of contact-distance restraints in the prediction of protein structures
with the UNRES force field. J. Comput. Chem., 2019,
https://doi.org/10.1002/jcc.25847 (Early View).
Original papers in non-ISI journals
- P. Arłukowicz, E. Arłukowicz, J. Kira, A. Liwo.
A user-friendly interface to computer programs for the determination of the
characteristics of equilibrium systems from physicochemical data.
TASK Quarterly, 1998, 2, 55-72.
- D.R. Ripoll, A. Liwo, C. Czaplewski.
The ECEPP package for conformational analysis of polypeptides.
TASK Quart., 1999, 3, 313-331.
- R. Ganzynkowicz, A. Liwo, W. Wiczk.
A fluorescence,
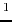 H NMR
spectroscopy and molecular dynamics study of the influence of
rotamer population on fluorescence decay of tyrosine,
phenylalanine and their derivatives.
TASK Quart., 2001, 5, 311-316.
H NMR
spectroscopy and molecular dynamics study of the influence of
rotamer population on fluorescence decay of tyrosine,
phenylalanine and their derivatives.
TASK Quart., 2001, 5, 311-316.
- D. Lapidus, S. Ventura, C. Czaplewski, A. Liwo, I. Liepina,
Molecular dynamics of amylin amyloid single
 -sheet.
Scientific Journal of Riga Technical University,
2011, 23, 49-55.
-sheet.
Scientific Journal of Riga Technical University,
2011, 23, 49-55.
- V. Duka, I. Bestel, C. Czaplewski, A. Liwo, I. Liepina,
Molecular modeling of single
 -sheet and the
-sheet and the  -sheet
stack of amyloid
-sheet
stack of amyloid  -protein 25 - 35.
Molecular dynamics of amylin amyloid single
-protein 25 - 35.
Molecular dynamics of amylin amyloid single  -sheet.
Scientific Journal of Riga Technical University,
2011, 23, 62-72.
-sheet.
Scientific Journal of Riga Technical University,
2011, 23, 62-72.
- M.A. Mozolewska, P. Krupa, B. Rasulev, A. Liwo, L. Leszczynski.
Preliminary studies of interactions between nanotubes and
toll-like receptors.
TASK Quarterly, 2014, 18, 351-455.
- E. Lubecka, A. K. Sieradzan, C. Czaplewski, P. Krupa, A. Liwo.
High performance computing with coarse grained model of biological
macromolecules.
Supercomputing Frontiers and Innovations, 2018, 5, 63-67.
Review articles in ISI journals
- M. Tarnowska, A. Liwo, F. Kasprzykowski, L. Łankiewicz, J. Ciarkowski.
Some aspects of the structure-activity relationships in oxytocin and
vasopressin.
Curr. Topics in Med. Chem., 1993, 1, 145-154.
- A. Liwo, S. Ołdziej, R. Kaźmierkiewicz, M. Groth, C. Czaplewski.
Design of a knowledge-based force field for off-lattice simulations of protein
structure.
Acta Biochim. Pol., 1997, 44, 527-548.
- H.A. Scheraga, J. Lee, J. Pillardy, Y.-J. Ye, A. Liwo, D.R. Ripoll.
Surmounting the Multiple-Minima Problem in Protein Folding.
J. Global Optimization, 1999, 15, 235-260.
- H.A. Scheraga, J. Pillardy, A. Liwo, J. Lee, C. Czaplewski, D.R. Ripoll,
W.J. Wedemeyer, Y.A. Arnautova.
Evolution of physics-based methodology for exploring the conformational energy
landscape of proteins.
J. Comput. Chem., 2002, 23, 28-34.
- H.A. Scheraga, A. Liwo, S. Ołdziej, C. Czaplewski, J. Pillardy, D. R. Ripoll,
J. A. Vila, R. Kazmierkiewicz, J. A. Saunders, Y. A. Arnautova, A.
Jagielska, M. Chinchio, M. Nanias.
The protein folding problem: global optimization of force fields.
Frontiers in Bioscience, 2004, 9, 3296-3323.
- J. Malicka, C. Czaplewski, M. Groth, W. Wiczk, S. Ołdziej, L. Łankiewicz,
J. Ciarkowski, A. Liwo.
Use of NMR and fluorescence spectroscopy as well as theoretical conformational
analysis in conformation-activity studies of cyclic enkephalin analogues.
Current Topics in Medicinal Chemistry, 2004, 4, 123-133.
- H.A. Scheraga,
 M. Khalili, A. Liwo.
Protein-folding dynamics: Overview of molecular simulation techniques.
Annu. Rev. Phys. Chem., 2007. 58, 57-83.
M. Khalili, A. Liwo.
Protein-folding dynamics: Overview of molecular simulation techniques.
Annu. Rev. Phys. Chem., 2007. 58, 57-83.
- A. Liwo, C. Czaplewski, S. Ołdziej, H.A. Scheraga.
 Computational techniques for efficient conformational sampling of proteins.
Curr. Opinion. Struct. Biol., 2008, 18, 134-139.
Computational techniques for efficient conformational sampling of proteins.
Curr. Opinion. Struct. Biol., 2008, 18, 134-139.
- A. Lewandowska, S. Ołdziej, A. Liwo, H.A. Scheraga.
 -hairpin-forming peptides; models of early stages of protein folding.
Biophys. Chem., 2010, 151, 1-9.
-hairpin-forming peptides; models of early stages of protein folding.
Biophys. Chem., 2010, 151, 1-9.
- A. Liwo, Y. He, H.A. Scheraga.
Coarse-grained force field: general folding theory.
Phys. Chem. Chem. Phys., 2011, 13, 16890-16901.
Review articles in non-ISI journals
- A. Liwo, M. Tarnowska, J. Ciarkowski, Z. Grzonka.
Application of Free-Wilson method in structure-activity correlation of
bioactive compounds based on the example of oxytocin and vasopressin analogs.
Wiad. Chem., 1994, 48, 805-815.
- P. Arłukowicz, E. Biernat, J. Ciarkowski, C. Czaplewski, M. Groth,
R. Kaźmierkiewicz, A. Liwo, K. Maksimiak, M. Nowacka, S. Ołdziej,
S. Rodziewicz, E. Woźniak.
Molecular simulations with high-performance computers.
TASK. Quarterly, 1997, 1, 21-37.
Book chapters
- S. Talluri, J.S.R. Murthy, A. Liwo, Structural Bioinformatics, in
``Fundamentals of Bioinformatics'' ed. I.A. Khan i A. Khanum, Ukaaz
Publications, Hyderabad, India, 2003, pp. 45-90.
- H.A. Scheraga, A. Liwo, S. Ołdziej, C. Czaplewski, J. Pillardy,
J. Lee, D.R. Ripoll, J.A. Vila, R. Kaźmierkiewicz, J.A. Saunders,
Y.A. Arnautova, K.D. Gibson, A. Jagielska, M. Khalili, M. Chinchio,
M. Nanias, Y.K. Kang, H. Schefroth, A. Ghosh, R. Elber, M. Makowski.
The Protein Folding Problem.
in ``New algorithms for macromolecular simulation -
Lecture Notes in Computational Science and Engineering'',
ed. B. Leimkuhler, C. Chipot, R. Elber, A. Laaksonen, A. Mark, T. Schlick,
C. Schütte, R. Skeel, Springer, Berlin, 2006.
- A. Liwo, C. Czaplewski, S. Ołdziej, A.V. Rojas, R. Kaźmierkiewicz,
M. Makowski, R.K. Murarka, H.A. Scheraga.
Simulation of protein structure and dynamics with the coarse-grained UNRES
force field. In: Coarse-Graining of Condensed Phase and Biomolecular
Systems., ed. G. Voth, Taylor & Francis, 2008, Chapter 8, pp. 107-122.
- D.R. Ripoll, A. Liwo, H.A. Scheraga.
Global Optimization in Protein Folding.
In Encyclopedia of Global Optimization,LE-TeX Jelonek,
Schmidt & Vöckler GbR, 2008, pp. 1391-1411.
- C. Czaplewski, A. Liwo, H.A. Scheraga.
Hydrophobic Interactions and Protein Folding.
In Thermodynamics of Amino Acid and Proein Solutions,
ed. J. Tsurko, W. Kunz, Transworld Research Network, Kerala, India, 2010,
pp. 1-18.
- C. Czaplewski, A. Liwo, M. Makowski, S. Ołdziej, H.A. Scheraga,
``Coarse-Grained Models of Proteins: Theory and Applications'', in A. Kolinski
(ed) "Multiscale Approaches to Protein Modeling", Springer, 2010.
- S. Ołdziej, C. Czaplewski, A. Liwo, J.A. Vila, H.A. Scheraga,
``Computation of Structure, Dynamics, and Thermodynamics of Proteins'',
in E. Engelman (ed) Comprehensive Biophysics, Vol. 1 Biophysical
Techniques for Structural Characterization of Macromolecules, ed. J. Dyson,
Oxford: Academic Press, pp. 494-513.
- A. Liwo, A. Niemi, X. Peng, A.K. Sieradzan,
A Novel Coarse-Grained Description of Protein Structure and
Folding by UNRES Force Field and Discrete Nonlinear
Schr
 inger Equation, in Frontiers in Computational Chemistry,
ed. by Zaheer-ul-Haq and J. D. Madura,
Vol. 1, pp. 257-289, Bentham Science Publishers, 2015.
inger Equation, in Frontiers in Computational Chemistry,
ed. by Zaheer-ul-Haq and J. D. Madura,
Vol. 1, pp. 257-289, Bentham Science Publishers, 2015.
- M. Baranowski, C. Czaplewski, E. Gołaś, Y. He, D. Jagieła, P. Krupa,
A. Liwo, G.G. Maisuradze, M. Makowski, M.A. Mozolewska, A. Niadzvedtski,
A.J. Niemi, S. Rackovsky, R. Ślusarz, A.K. Sieradzan, S. Ołdziej,
T. Wirecki, Y. Yin, B. Zaborowski, H.A. Scheraga, ``Microscopic physics-based
models of proteins and nucleic acids: UNRES and NARES'', in
Coarse-Grained Models of Biomolecules, ed. by Garegin A. Papoian,
CRC Press, ISBN 9781466576063, CAT# K16726, 2017.
- A. Liwo, C. Czaplewski, S. Ołdziej, B. Zaborowski, D. Jagieła, J. Lee.
Chemoinformatics Methods for Studying Biomolecules.
J. Leszczynski et al. (eds.), Handbook of Computational Chemistry,
Part IV: Biomolecules, Jerzy Leszczynski, Anna Kaczmarek-Kedziera, Tomasz Puzyn, Manthos G. Papadopoulos, Heribert Reis, Manoj K. Shukla, eds., pp 2183-2199, DOI 10.1007/978-94-007-6169-8_56-1, Springer, 2017.
- E.I. Gołaś, M.A. Mozolewska, P. Krupa, C. Czaplewski, H.A. Scheraga,
A. Liwo,
Use of Coarse-Grained and All-Atom Molecular Dynamics to Study Hsp70 and Hsp40 Chaperone Action,
In: Role of Molecular Chaperones in Structural Folding, Biological Functions, and Drug Interactions of Client Proteins
Book Series: Frontiers in Structural Biology, Mario D. Galigniana (Ed.), Vol. 1, pp. 23-46, Bentham Science, 2018,
eISBN: 978-1-68108-615-6, 2018
ISBN: 978-1-68108-616-3,
ISSN: 2589-4366 (Print),
ISSN: 2589-4374 (Online).
- A. Liwo, A. K. Sieradzan, C. Czaplewski.
Formation of Secondary and Supersecondary Structure of Proteins
as a Result of Coupling Between Local and Backbone-Electrostatic
Interactions: A View Through Cluster-Cumulant Scope.
In: Protein Supersecondary Structures. Alexander E. Kister (Ed.),
pp. 133-146,
Humana Press, Springer, N.Y., 233 Spring Street, New York, NY 10013, U.S.A.,
2-10.
pp. 133-146.
Books edited
- Computational methods to study the structure and dynamics of biomolecules
and biomolecular processes, ed. by A. Liwo,
Springer, Heidelberg, 2014, ISBN 978-3-642-28553-0.
Papers in refereed conference proceedings
- Z. Grzonka, L. Łankiewicz, F. Kasprzykowski, A. Liwo, P. Melin.
Structure-activity relationships of vasopressin: synthesis of new vasopressin
analogues. In Peptides 1986 Theodoropoulos, ed., Walter de Gruyter 1987,
pp. 493-496.
- F. Kasprzykowski, P. Kania, S. Ołdziej, A. Liwo, M. Tarnowska,
Z. Grzonka, M. Abrahamson, I. Olafsson, A. Grubb and J. Trojnar. Inhibition of
cysteine proteinases by a series of peptidyl-diazomethanes structurally
related to the binding center of cystatins. In Peptides 1990,
eds. E. Ginolt, D. Andrew, ESCOM, Leiden 1991, pp. 799-800.
- Z. Grzonka, E. Gwizdała, F. Kasprzykowski, A. Liwo,
L. Łankiewicz, L. Łubkowska, S.G. Galaktionov, V.M. Tseitsin.
Studies on interaction of neurohypophyseal hormones with lipids.
In Peptides 1990, eds. E. Ginolt, D. Andrew, ESCOM, Leiden 1991,
pp. 488-489
- S. Ołdziej, F. Kasprzykowski, L. Łankiewicz, P. Kania,
A. Liwo and Z. Grzonka. Proposed mechanism of interaction of type II cystatins
with papain.
In Peptides 1991, eds. J.A. Smith, J.E.Rivier, ESCOM, Leiden 1991,
pp. 793-794.
- M. Tarnowska, A. Liwo, Z. Grzonka, A. Tempczyk.
Modified Free-Wilson analysis of the neurohypophyseal hormone
analogs I: Analogs of vasopressin-like activity.
In QSAR in Design of Bioactive Compounds, Vol. 2, ed. M. Kuchar,
J.R. Prous Science Publishers: Barcelona 1991, pp. 361-397.
- M. Tarnowska, A. Liwo, Z. Grzonka, A. Tempczyk.
Modified Free-Wilson analysis of the neurohypophyseal hormone
analogs II: Analogs of oxytocin-like activity.
In QSAR in Design of Bioactive Compounds, Vol. 2, ed. M. Kuchar,
J.R. Prous Science Publishers: Barcelona 1991, pp. 399-436.
- J. Ciarkowski, S. Ołdziej, M. Nowacka, A. Liwo, F.M.F. Chen,
and L. Benointon.
Mixed anhydride reactivity by means of the molecular orbital AM1 and PM3
methods.
Peptides 1994, ed. H.L.S. Maia, ESCOM, Leiden 1995, pp. 213-214.
- M.D. Shenderovich, K.E. Kövér, S. Wilke, M. Romanowski,
A. Liwo, L. Łankiewicz, E. Gwizdała, J. Ciarkowski, V.J. Hruby.
Conformational analysis of potent bicyclic antagonists of oxytocin: NMR,
CD and molecular dynamics study. Peptides 1995, (Proc. 14th APS),
Kaumaya, P.T.P. and Hodges, R.S., Eds., Mayflower Scientific Ltd.,
England 1996, 499-500.
- B. Micewicz, A. Liwo, L. Łankiewicz, W. Wiczk.
Mechanism of tyrosine fluorescence quenching by acrylamide in model peptides.
Peptides 1996, Ramage & Epton, (eds.), 1998, 649-650.
- S. Rodziewicz, A. Lesner, K. Rolka, A. Łęgowska, C. Czaplewski,
A. Liwo, R. Patacchini, L. Quartara.
Conformational studies of scyliorhinin II analogues - tachykinin peptides
using NMR spectroscopy and Monte Carlo global conformational analysis.
Peptides 1998, S. Bajusz and F. Hudecz (eds.), Akademiai Kiadó,
Budapest, 1999, 334-335.
- J. Malicka, C. Czaplewski, M. Groth, A. Kubicki, P. Arłukowicz,
W. Wiczk, A. Liwo.
Studies of inter-pharmacophore distance in cyclic enkephalin analogues
by fluorescence spectroscopy and molecular dynamics.
Peptides 1998, S. Bajusz and F. Hudecz (eds.), Akademiai Kiadó,
Budapest, 1999, 368-369.
- M. Groth, J. Malicka, C. Czaplewski, W. Wiczk, A. Liwo.
Conformational studies of cyclic enkephalin analogues with L- or D-proline
in position 3 by NMR spectroscopy and global conformational analysis.
Peptides 1998, S. Bajusz and F. Hudecz (eds.), Akademiai Kiadó,
Budapest, 1999, 370-371.
- S. Rodziewicz, I. Wirkus-Romanowska, M. Ciurak, H. Miecznikowska, C. Czaplewski,
A. Liwo, K. Rolka, G. Kupryszewski.
Cyclic analogues of proline-rich protein fragment, synthesis and conformational
studies.
Peptides 1998, S. Bajusz and F. Hudecz (eds.), Akademiai Kiadó,
Budapest, 1999, 448-449.
- A. Liwo, J. Pillardy, C. Czaplewski, J. Lee, D.R. Ripoll, M. Groth,
S. Rodziewicz-Motowidlo, R. Kazmierkiewicz, R.J. Wawak, S. Ołdziej,
H.A. Scheraga,
UNRES - a united-residue force field for energy-based
prediction of protein structure - origin and significance of multibody
terms,
RECOMB 2000, Proceedings of the Fourth Annual International
Conference on Computational Molecular Biology, R. Shamir, S. Miyano, S.
Istrail, P. Pevzner, M. Waterman (Eds.), April 8-11, Tokyo, Japan,
ACM, New York, pp. 193-200.
- R. Ganzynkowicz, A. Rzeska, A. Liwo, W. Wiczk.
Influence of a rotamer population on fluorescence decay of the tyrosine
derivatives studied by fluorescence,
 HNMR spectroscopy, and theoretical
calculation.
Peptides 2000, J. Martinez and J.-A. Fehrentz (eds.), EDK, Paris,
2001, 411-412.
HNMR spectroscopy, and theoretical
calculation.
Peptides 2000, J. Martinez and J.-A. Fehrentz (eds.), EDK, Paris,
2001, 411-412.
- S. Rodziewicz-Motowidło, K. Brzozowski, A. Łęgowska, A. Liwo, K. Rolka,
J. Silbering, M. Smoluch.
Conformational studies of neuropeptide
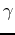 using CD, 2D NMR spectroscopy
and theoretical analysis.
Peptides 2000, J. Martinez and J.-A. Fehrentz (eds.), EDK, Paris,
2001, 459-460.
using CD, 2D NMR spectroscopy
and theoretical analysis.
Peptides 2000, J. Martinez and J.-A. Fehrentz (eds.), EDK, Paris,
2001, 459-460.
- I. Liepiņa, C. Czaplewski, P. Janmey, A. Liwo.
Molecular dynamics of the gelsolin-derived peptide binding to a
PIP
 -containing lipid bilayer.
Peptides 2000, J. Martinez and J.-A. Fehrentz (eds.), EDK, Paris,
2001, 987-988.
-containing lipid bilayer.
Peptides 2000, J. Martinez and J.-A. Fehrentz (eds.), EDK, Paris,
2001, 987-988.
- I. Liepina, P. Janmey, C. Czaplewski, A. Liwo.
Molecular dynamics of the caspase-3 cleaved N-half of gelsolin.
Biopolymers, 2003, 71, 331.
- J. Ciarkowski, P. Drabik, Z. Grzonka, V. Hruby, A. Liwo, S. Ołdziej,
E. Politowska, M.D. Shenderovich, M. Witt.
From modeling neurohypophyseal hormones to signaling mechanisms in GCPRs.
Peptides 2004, M. Flegel, M. Fridkin, C. Gilon, J. Slaninova (eds.),
Keynes, Geneva, 2005, 125-127.
- I. Liepiņa, P. Janmey, C. Czaplewski, A. Liwo.
Towards gelsolin amyloid formation.
Peptides 2004, M. Flegel, M. Fridkin, C. Gilon, J. Slaninova (eds.),
Keynes, Geneva, 2005, 740-741.
- J. Łągiewka, S. Rodziewicz-Motowidło, R. Sosnowska, K. Stachowiak,
S. Ołdziej, A. Liwo, Z. Grzonka.
Investigation of the hCC dimerization mechanism - CD and NMR structural
stability studies of the
 2-L1-
2-L1- 3 fragment.
Peptides 2004, M. Flegel, M. Fridkin, C. Gilon, J. Slaninova (eds.),
Keynes, Geneva, 2005, 1029-1030.
3 fragment.
Peptides 2004, M. Flegel, M. Fridkin, C. Gilon, J. Slaninova (eds.),
Keynes, Geneva, 2005, 1029-1030.
- H.A. Scheraga, A. Liwo, St. Ołdziej, C. Czaplewski, M. Khalili, J.A. Vila,
D.R. Ripoll.
The two aspects of the protein folding problem.
NIC series, NIC Workshop 2006, From Computational Physics to System Biology,
Ulrich H.E. Hansmann, Jan Meinke, Sandipan Mohanty, Olav Zimmermann
(Editors), Volume 24, pp. 37-44.
- C. Czaplewski, S. Kalinowski, S. Ołdziej, A. Liwo, H.A. Scheraga.
Multiplexed replica exchange molecular dynamics with the UNRES force field
as an effective method for exploring conformational energy landscape of
proteins.
NIC series, NIC Workshop 2006, From Computational Physics to System Biology,
Ulrich H.E. Hansmann, Jan Meinke, Sandipan Mohanty, Olav Zimmermann
(Editors), Volume 24, pp. 63-66.
- I. Liepiņa, S. Ventura, C. Czaplewski, A. Liwo.
Molecular dynamics study of amyloid formation of two Abl-SH3 domain
peptides.
NIC series, NIC Workshop 2006, From Computational Physics to System Biology,
Ulrich H.E. Hansmann, Jan Meinke, Sandipan Mohanty, Olav Zimmermann
(Editors), Volume 24, pp. 103-106.
- A. Liwo, M. Khalili, C. Czaplewski, S. Kalinowski, S. Ołdziej, H.A. Scheraga.
Optimization of the united-residue UNRES force field for Langevin dynamics
simulations.
NIC series, NIC Workshop 2006, From Computational Physics to System Biology,
Ulrich H.E. Hansmann, Jan Meinke, Sandipan Mohanty, Olav Zimmermann
(Editors), Volume 24, pp. 107-109.
- U. Kozłowska, K. Wachucik, A. Liwo, H.A. Scheraga.
Determination of short-range potentials for physics-based protein structure
prediction.
NIC series, NIC Workshop 2006, From Computational Physics to System Biology,
Ulrich H.E. Hansmann, Jan Meinke, Sandipan Mohanty, Olav Zimmermann
(Editors), Volume 24, pp. 169-172.
- F. Rakowski, P. Grochowski, B. Lesyng, A. Liwo, H.A. Scheraga.
Symplectic algorithms of molecular dynamics. An application to the
united-residue model.
NIC series, NIC Workshop 2006, From Computational Physics to System Biology,
Ulrich H.E. Hansmann, Jan Meinke, Sandipan Mohanty, Olav Zimmermann
(Editors), Volume 24, pp. 181-184.
- M. Makowski, A. Liwo, K. Maksimiak, J. Makowska, L. Chmurzyński,
H.A. Scheraga.
Simple physics-based analytical formulas for the potentials of mean force
for the interaction for amino-acid side chains in water. Tests with simple
spherical systems.
NIC series, NIC Workshop 2006, From Computational Physics to System Biology,
Ulrich H.E. Hansmann, Jan Meinke, Sandipan Mohanty, Olav Zimmermann
(Editors), Volume 24, pp. 111-114.
- A. Liwo, C. Czaplewski, S. Ołdziej, M. Chinchio, A.V. Rojas, M. Khalili,
M. Makowski, S. Kalinowski, U. Kozłowska, R.K. Murarka, H.A. Scheraga.
Mesoscopic dynamics with the UNRES force field - a tool for studying
the kinetics and thermodynamics of protein folding.
NIC series, NIC Workshop 2007, From Computational Physics to System Biology,
Ulrich H.E. Hansmann, Jan Meinke, Sandipan Mohanty, Olav Zimmermann
(Editors), Volume 36, pp. 31-36.
- I. Liepiņa, S. Ventura, C. Czaplewski, A. Liwo.
Multiple
 -sheet molecular dynamics of two Abl-SH3 domain peptides.
NIC series, NIC Workshop 2007, From Computational Physics to System Biology,
Ulrich H.E. Hansmann, Jan Meinke, Sandipan Mohanty, Olav Zimmermann
(Editors), Volume 36, pp. 215-218.
-sheet molecular dynamics of two Abl-SH3 domain peptides.
NIC series, NIC Workshop 2007, From Computational Physics to System Biology,
Ulrich H.E. Hansmann, Jan Meinke, Sandipan Mohanty, Olav Zimmermann
(Editors), Volume 36, pp. 215-218.
- M. Makowski, E. Sobolewski, A. Liwo, J. Makowska, L. Chmurzyński, H.A.
Scheraga.
Determination of potentials of mean force dependent on orientation
in hydrophobic systems modeling side-chains of biomolecules.
Peptides 2006, K. Rolka, P. Rekowski and J. Silberring (eds),
Kenes, Geneva 2007, 60-61.
- J. Makowska, K. Bagińska, M. Makowski, A. Liwo, L. Chmurzyński.
Evaluation of two theoretical methods to estimate potentiometric-titration
curves of peptides: comparison with experiment.
Peptides 2006, K. Rolka, P. Rekowski and J. Silberring (eds),
Kenes, Geneva 2007, 62-63.
- I. Liepiņa, S. Ventura, C. Czaplewski, A. Liwo.
Molecular dynamics study of two Abl-SH3 domain peptides towards amyloid
formation.
Peptides 2006, K. Rolka, P. Rekowski and J. Silberring (eds),
Kenes, Geneva 2007, 782-783.
- S. Rodziewicz-Motowidło, A. Skwierawska, S. Ołdziej, W. Wiczk, A. Liwo,
H.A. Scheraga, Conformational studies of a short peptide corresponding to the
C-terminal
 -hairpin structure of the B1 domain of protein G.
Peptides 2006, K. Rolka, P. Rekowski and J. Silberring (eds),
Kenes, Geneva 2007, 794-795.
-hairpin structure of the B1 domain of protein G.
Peptides 2006, K. Rolka, P. Rekowski and J. Silberring (eds),
Kenes, Geneva 2007, 794-795.
- A. Liwo, C. Czaplewski, S. Ołdziej, U. Kozłowska, M. Makowski, S. Kalinowski,
R. Kaźmierkiewicz, H. Shen, G. Maisuradze, H.A. Scheraga,
Optimization of a physics-based united-residue force field (UNRES) for protein
folding simulations, NIC Series, NIC Symposium 2008, eds. G. M
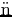 ster, D. Wolf
and M. Kremer, 2008, 39, 63-70.
ster, D. Wolf
and M. Kremer, 2008, 39, 63-70.
- I. Liepina, S. Ventura, C. Czaplewski, A. Liwo.
Dynamics study on single and multiple
 -sheets.
Peptides for Youth, Book Series: Advances in Experimental Medicine and Biology,
Vol. 611, pp. 293-294.
-sheets.
Peptides for Youth, Book Series: Advances in Experimental Medicine and Biology,
Vol. 611, pp. 293-294.
- S. Ołdziej, C. Czaplewski, A. Liwo, H.A. Scheraga,
Towards Temperature Dependent Coarse-grained Potential of Side-chain
Interactions for Protein Folding Simulations, BIBE, pp.263-266, 2010
IEEE International Conference on Bioinformatics and Bioengineering, 2010
Józef Adam Liwo
Invited presentations
- A. Liwo.
The role of hydrophobic residue packing in protein folding.
8th Conference of Young Scientists on Organic, Bioorganic Chemistry,
Riga, Latvia, 2-9 November 1991.
- Application of the Free-Wilson method in structure-activity correlation
of bioactive compounds on the example of oxytocin and vasopressin analogs.
Theoretical and Experimental Aspects of Molecular Structure, Karpacz,
Poland, June 5-10, 1994
- Prediction of protein structure using a mean-field united-residue potential
determined from protein crystal data. 3-rd Conference Computers in Chemistry
'94, Wrocław, Poland, June 23-26, 1994.
- Role of singlet-oxygen binding to anthraquinones in the peroxidating activity
of antitumor anthraquinone drugs. 5th International Symposium on Molecular
Aspects of Chemotherapy, Gdansk, Poland, August 21-24, 1995
- A knowledge-based united-residue force field for off-lattice calculations
of protein structure that recognizes native folds.
International Symposium on Theoretical, Experimental Aspects of Protein
Folding, San Luis, Argentina, June 17-21, 1996
- Energy-based prediction of protein structure with the UNRES force field,
First KIAS Conference on Protein Structure and Function: Protein Folding in
Post-Genome Era, Korea Institute for Advanced Study, Seoul, Korea,
November 28 - November 30, 2001.
- Design of hierarchical caldera-like potential-energy functions for
energy-based prediction of protein structure and simulation of protein
folding:application to the UNRES force field. Third KIAS Conference on
Protein Structure and Function: Folding Mechanism, Proteomics, and
Bioinformatics, Seoul, Korea, 29.09-1.10, 2003, 2003,
- Ab initio simulations of protein folding pathways by molecular dynamics
with the united-residue (UNRES) model of polypeptide chains.
30th FEBS Congress and 9th IUBMB Conference, Budapest, Hungary, 2005;
FEBS Journal., 2005, 272 (Suppl. 1) 359.
- Optimization of a mesoscopic force field for simulation of protein folding
pathways. Workshop on
Structure and Function of Biomolecules May 13-15, 2006, B
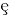 dlewo
near Poznań, Poland.
dlewo
near Poznań, Poland.
- Mesoscopic dynamics with the UNRES force field - a tool for studying the
kinetics and thermodynamics of protein folding.
NIC Workshop 2007, From Computational Biology to System Biology,
Jülich, Germany, May 2-4, 2007.
- Prediction of structure and simulation of dynamics of protein folding with
the mesoscopic UNRES force field, Modelling and Design of Molecular Materials.
Piechowice, Poland, June 23-28, 2008.
- Towards simulations of structure and dynamics of large proteins with the
UNRES force field.
The 8th KIAS - Yonsei Conference on Protein Structure and Function, Seoul,
Korea, October 9-11, 2008.
- Use of the UNRES force field and massively parallel computers in millisecond
scale simulations of protein
dynamics. Workshop on Bioinformatics (BIT09), Toruń, Poland May 23-25, 2009.
- The nature of the conformational ensemble at the transition temperature:
insights from simulations and experiment.
Third Korea-Japan Seminars on Biomolecular Sciences: Experiments and
Simulations, Hotel Lotte, Jeju, Korea, February 26, March 1, 2011.
- From atomistic simulations to network description of biological systems,
Workshop ``From Computational Biophysics
to Systems Biology'' 2011 (CBSB11), Juelich, Germany, July 20-22, 2011.
- Construction and application of coarse-grained force fields for
biomacromolecules, Multipole Approaches to
Structural Biology, Warsaw, Poland, November 16-19, 2011.
- Coarse-grained models for proteins. Nordita Workshop
``Dynamics of Biomolecular Processes: From Atomistic
Representations to Coarse-Grained Models'', Stockholm, Sweden,
February 27 - March 23, 2012; a series of 4
invited lectures.
- Coarse grained description of biomolecular systems. 10th Workshop on
Bioinformatics and 5th Symposium of the
Polish Bioinformatics Society, Gdańsk, Poland, May 25-26, 2012.
- A simple coarse-grained model of nucleic acids reveals the essential
role of mean-field dipole-dipole interactions between nucleic-acid bases
and double-helix formation.
Fifth Korea-Japan Seminars on Biomolecular Sciences:
Experiments and Simulations.
High1 Resort, Republic of Korea, February 24-16, 2013.
- Mean field dipole-dipole interactions as essential factors in the formation
of biomolecular architectures.
Biomolecules and Nanostructures 4,
Pułtusk, Poland, May 15-19, 2013.
- Essential role of mean-field electrostatic interactions in free
modeling of protein and nucleic acid structures at coarse-grained level.
Protein & RNA Structure Prediction Conference 2013,
Occidental Grand Xcaret, Playa del Carmen Mexico, December 1-5, 2013.
- A maximum-likelihood approach to force-field calibration.
Protein Folding Conference 2014, Grand Palladium Resort & Spa, Bavaro
Hotel Punta Cana, Dominican Republic, July 16-19, 2014.
- The first Nobel Prize in computational chemistry. An invited lecture
presented during the quarterly Colloquium at the Inistitute of Physics,
Polish Academy of Sciences, Warsaw, Poland, February 25, 2014.
- Maximum-likelihood calibration of force fields.
1st Korean-Polish Conference on Protein Folding: Theoretical and
Experimental Approaches, Seoul, Rep. of Korea, May 24-28, 2015.
- A rigorous approach to the derivation of analytical potentials in physics-based
coarse-grained force fields. 3rd International Workshop on Theoretical and Computational
Physics, Complex Systems and Interdisciplinary Physics, Da Lat, Vietnam,
July 27-30, 2015.
- A unified coarse-grained model for biomolecular simulations. 2015 Workshop of Wuhan
Center of Physical Biology, Wuhan, P.R. China, September 7, 2015.
- The UNRES coarse-grained model of polypeptide chains: theory and applications.
Invited lecture presented at the Department of Physics, Beijing Institute of Technology,
Beijing, P.R. China, September 17, 2015.
- A rigorous approach to derive analytical expressions for the effective energy
terms in coarse-grained force fields. 3rd International Conference on Protein and
RNA Structure Prediction Punta Cana, Dominican Republic, Dec 14-18, 2015.
- How do the local and long-range interactions encode the three-dimensional structures
of biological macromolecules: a coarse-grained perspective.
6th Visegrad Symposium on Structural Systems Biology, Warsaw, Poland, 19-21 June, 2016.
- The Unified Coarse-Grained Model for large-scale simulations of biological macromolecules.
The 16th KIAS Conference on Protein Structure and Function, Seoul, Republic of Korea,
September 22 - September 24, 2016.
- A general method for the derivation of effective energy expressions in coarse-grained force fields.
3rd Korean-Polish Conference on Protein Folding: Theoretical and Experimental Approaches, Seoul, Republic of Korea, February 4 – 9, 2017
- Geometry-consistent expressions for energy terms in coarse-grained force fields for (bio)polymers with UNRES as an example.
Workshop on Physics and Biology of Proteins, Natal, Brazil, June 12-30, 2017
- Implementation of geometry-consistent local and correlation potentials �in the UNRES force field.
Biomolecules and Nanostructures 6, Podlesice, Poland, May 10-14, 2017
- Physics-based scale- and geometry-consistent coarse-grained potentials.
26th Conference on Current Trends in Computational Chemistry (2018 CCTCC)
Jackson, MS (USA), 9-10 November 2018.
- Systematic design of physics-based scale-consistent coarse-grained potentials
for the simulations of biomolecules and nanostructures,
Bio-, chem-, and nanoinformatics approaches to study bionano interface
Dublin, Ireland, May 23-25, 2018.
- Data-assisted prediction of protein structures with the coarse-grained UNRES force field.
CASP13 Meeting, Iberostar Paraiso Maya, Mexico, December 1-4, 2018.
- Using the UNRES server and the standalone UNRES package in SAXS-data-assisted
modeling of protein structure.
simSAS 2019, Grenoble France, April 8-11/12, 2019